Visualize differences between treatments
dat_shoot_alive <- dat_shoot %>%
drop_na(barcode) %>%
filter(shoot > 0) %>%
drop_na(shoot) %>%
group_by(barcode, year) %>%
mutate(shoot_alive = if_else(any(alive == 0), 0, 1)) %>% # filter out plants ever considered dead at any point in a year
ungroup() %>%
filter(shoot_alive == 1) %>%
select(-alive, -shoot_alive)
df_param_all <- calc_empirical_parameters(df = dat_shoot_alive, path = "alldata/intermediate/", log = T, overwrite = F) %>%
tidy_species_name()df_param_diff <- calc_parameter_diff(df_param_all)
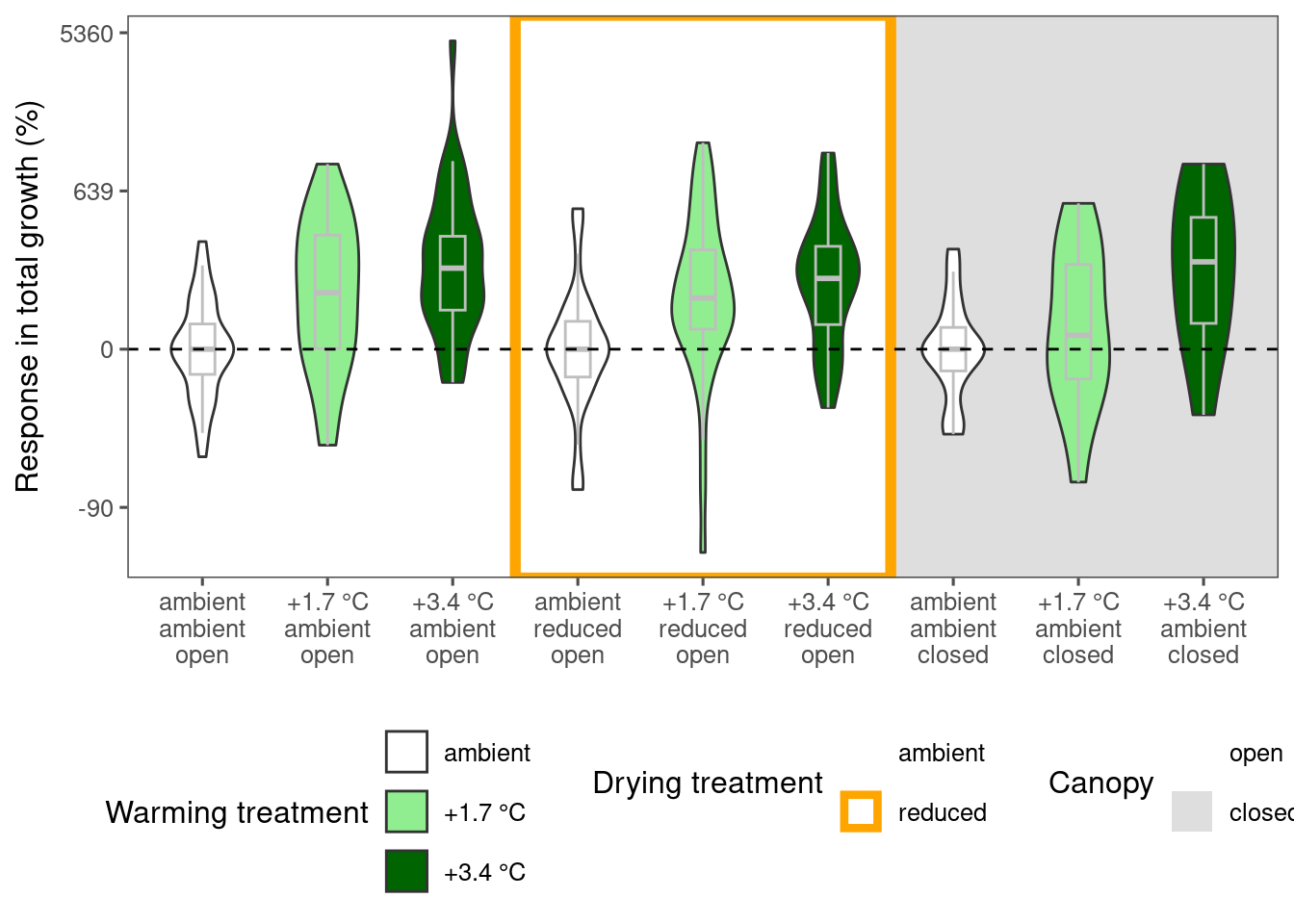
plot_metric_difference(
dat_diff = df_param_diff %>%
filter(
species %in% c("acesa"),
param %in% c("total growth")
), type = "shoot", x_label_short = F, simple = F
)
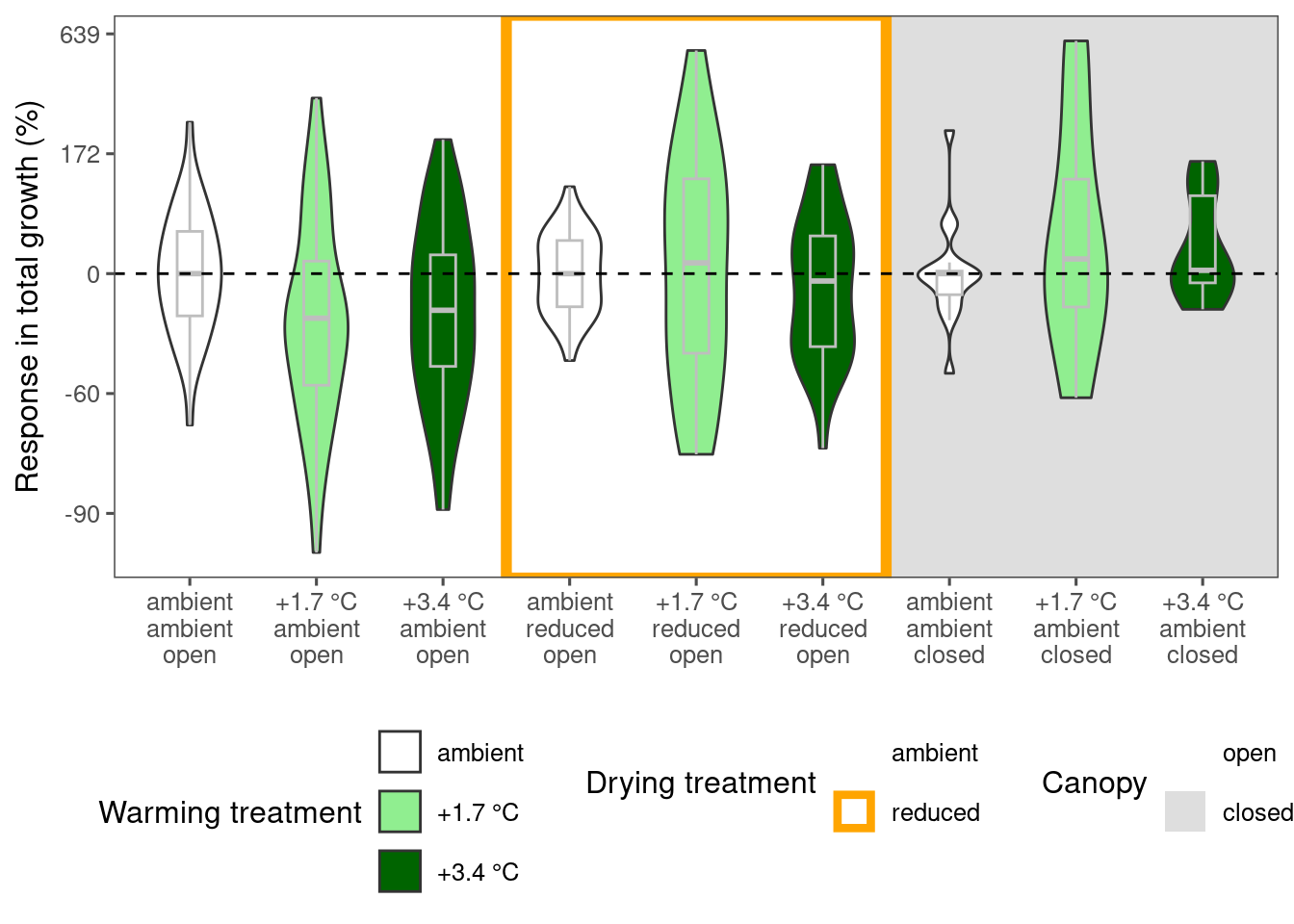
plot_metric_difference(
dat_diff = df_param_diff %>%
filter(
species %in% c("picgl"),
param %in% c("total growth")
), type = "shoot", x_label_short = F, simple = F
)
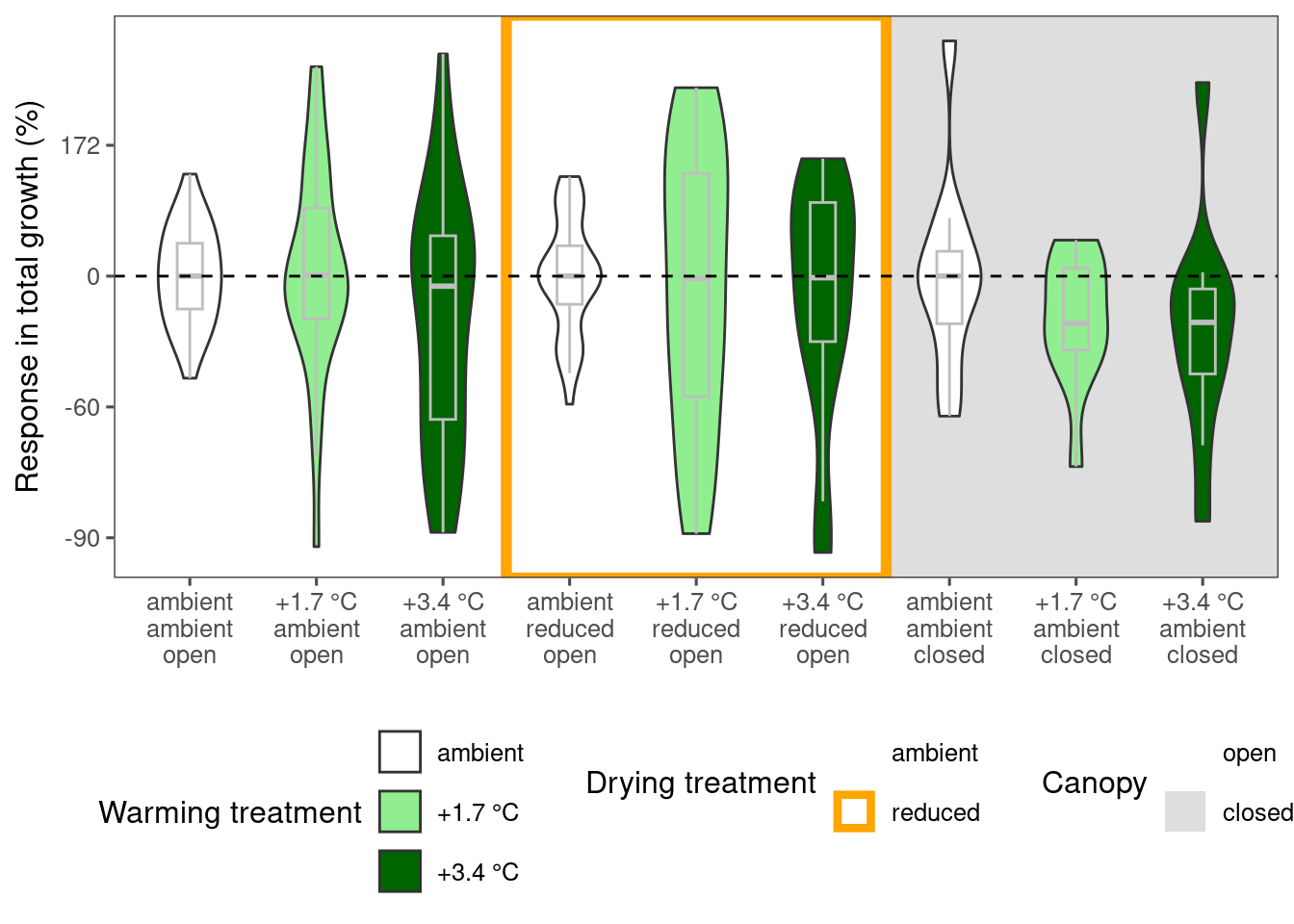
plot_metric_difference(
dat_diff = df_param_diff %>%
filter(
species %in% c("abiba"),
param %in% c("total growth")
), type = "shoot", x_label_short = F, simple = F
)
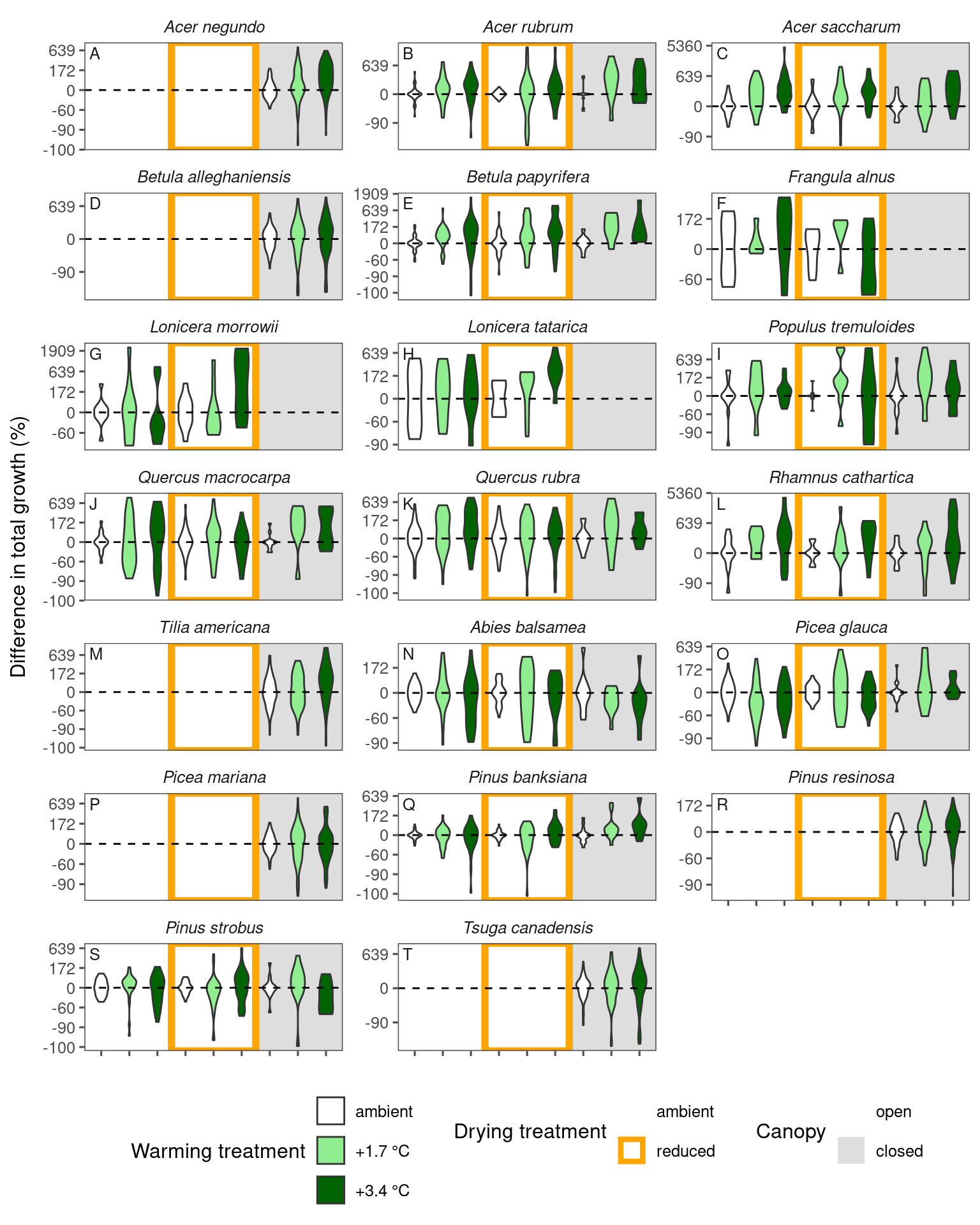
plot_metric_difference(
dat_diff = df_param_diff %>%
filter(param %in% c("total growth")), type = "shoot", x_label_short = T, simple = T
)
Visualize observational trends
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1, group_vars = c("site", "block", "plot", "canopy", "heat", "heat_name", "water", "water_name", "year"))plot_observational_trend(df_param_all %>% filter(species %in% c("acesa"), param == "total growth"), dat_climate_spring, type = "shoot")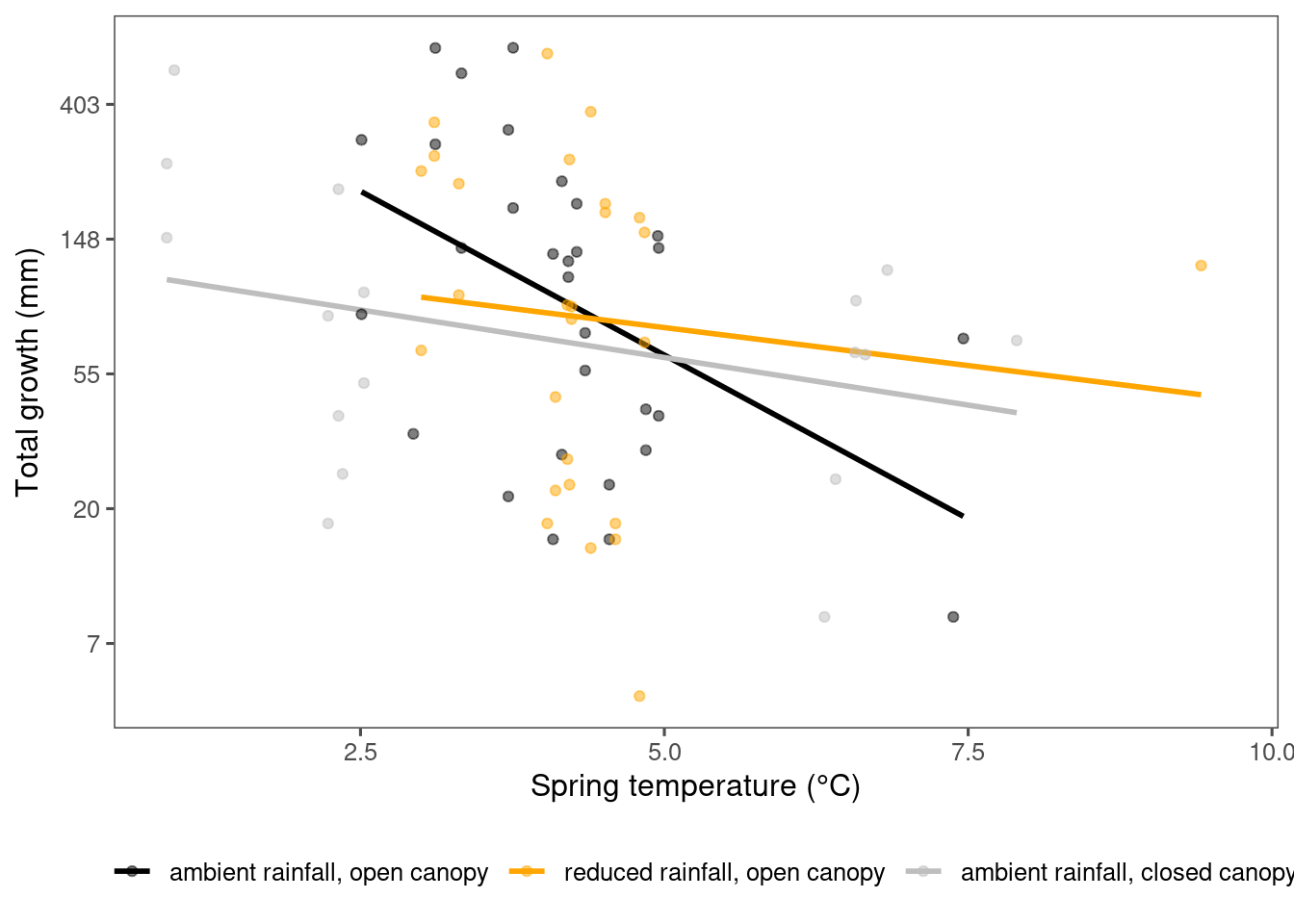
plot_observational_trend(df_param_all %>% filter(species %in% c("picgl"), param == "total growth"), dat_climate_spring, type = "shoot")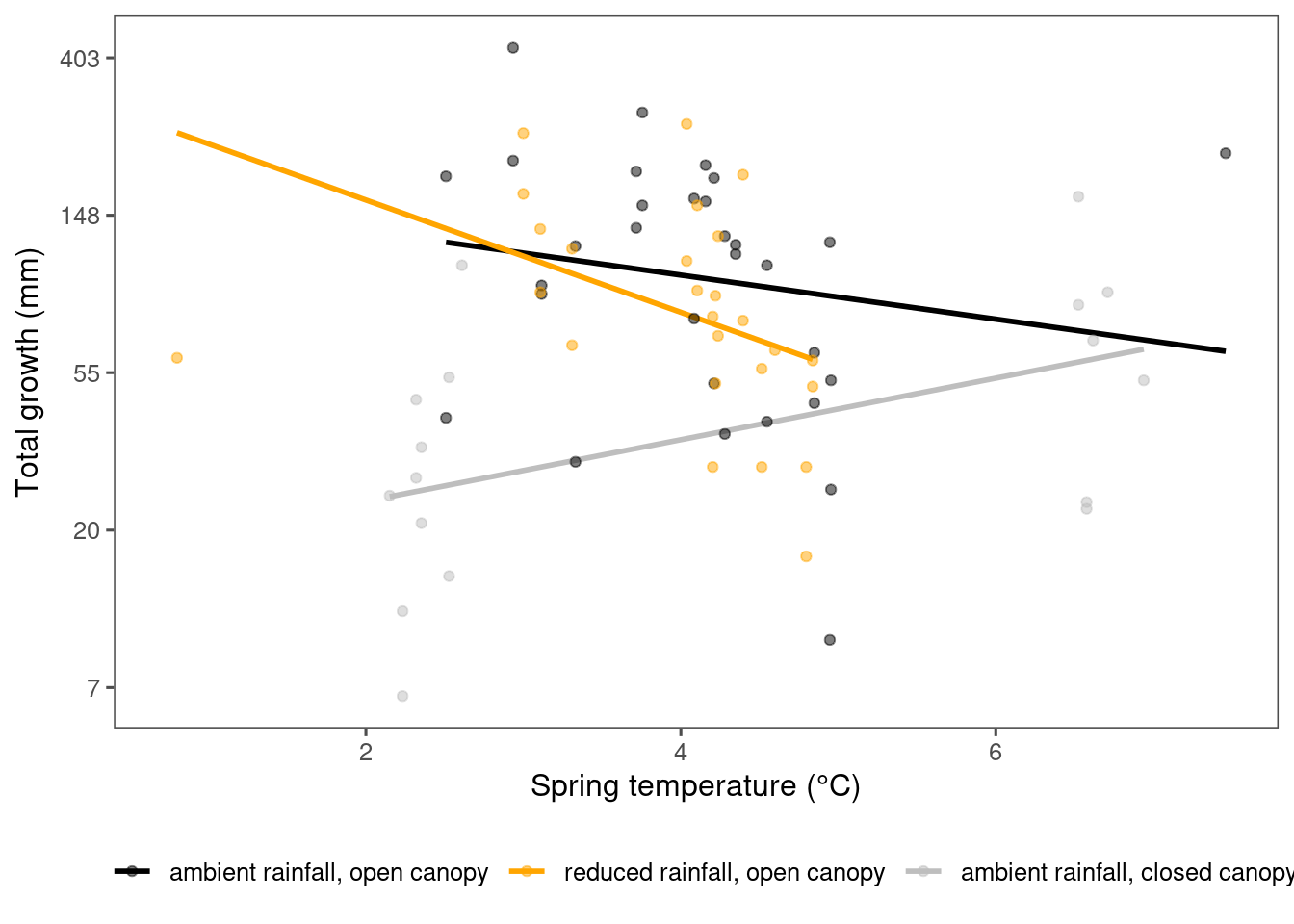
plot_observational_trend(df_param_all %>% filter(species %in% c("abiba"), param == "total growth"), dat_climate_spring, type = "shoot")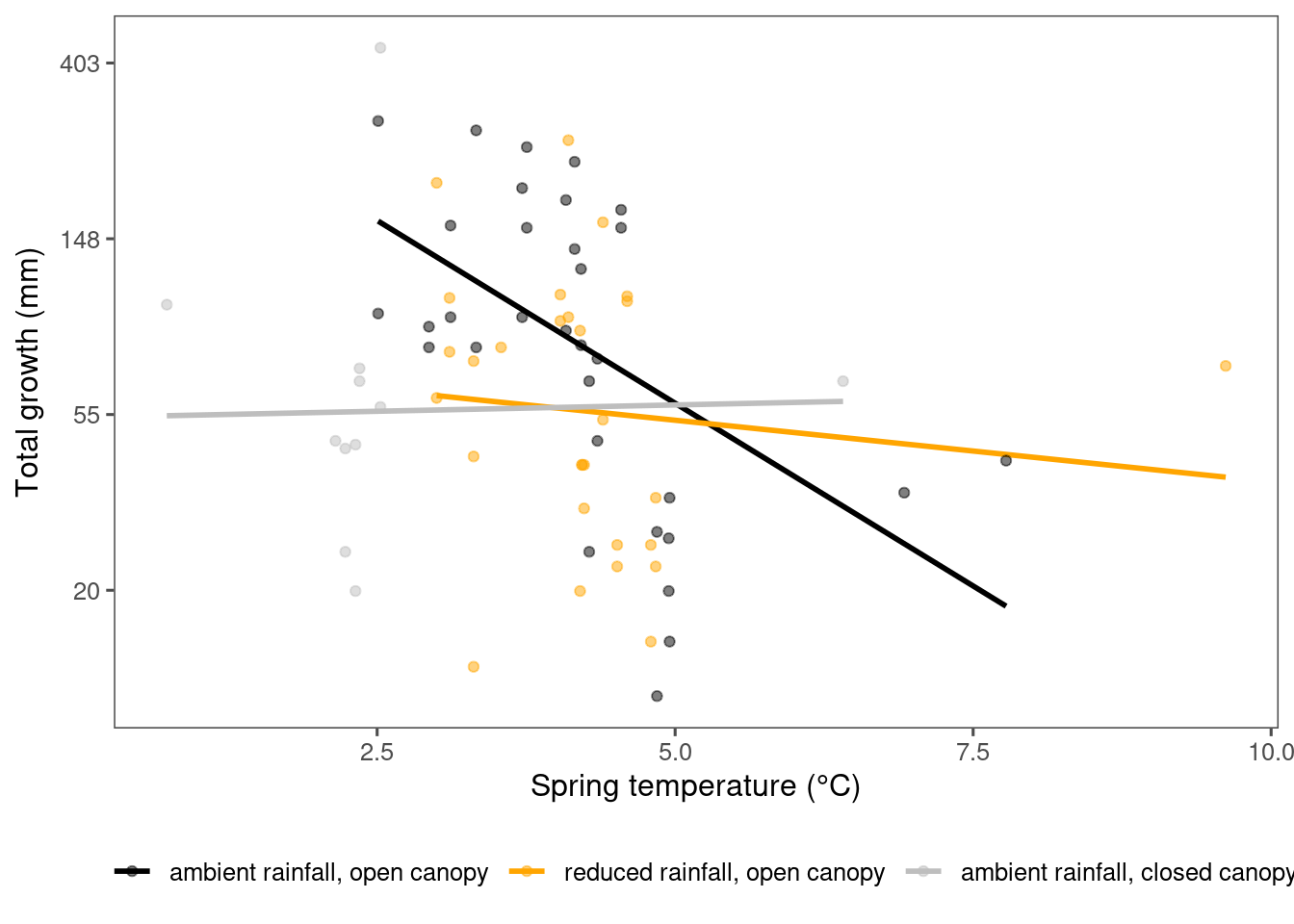
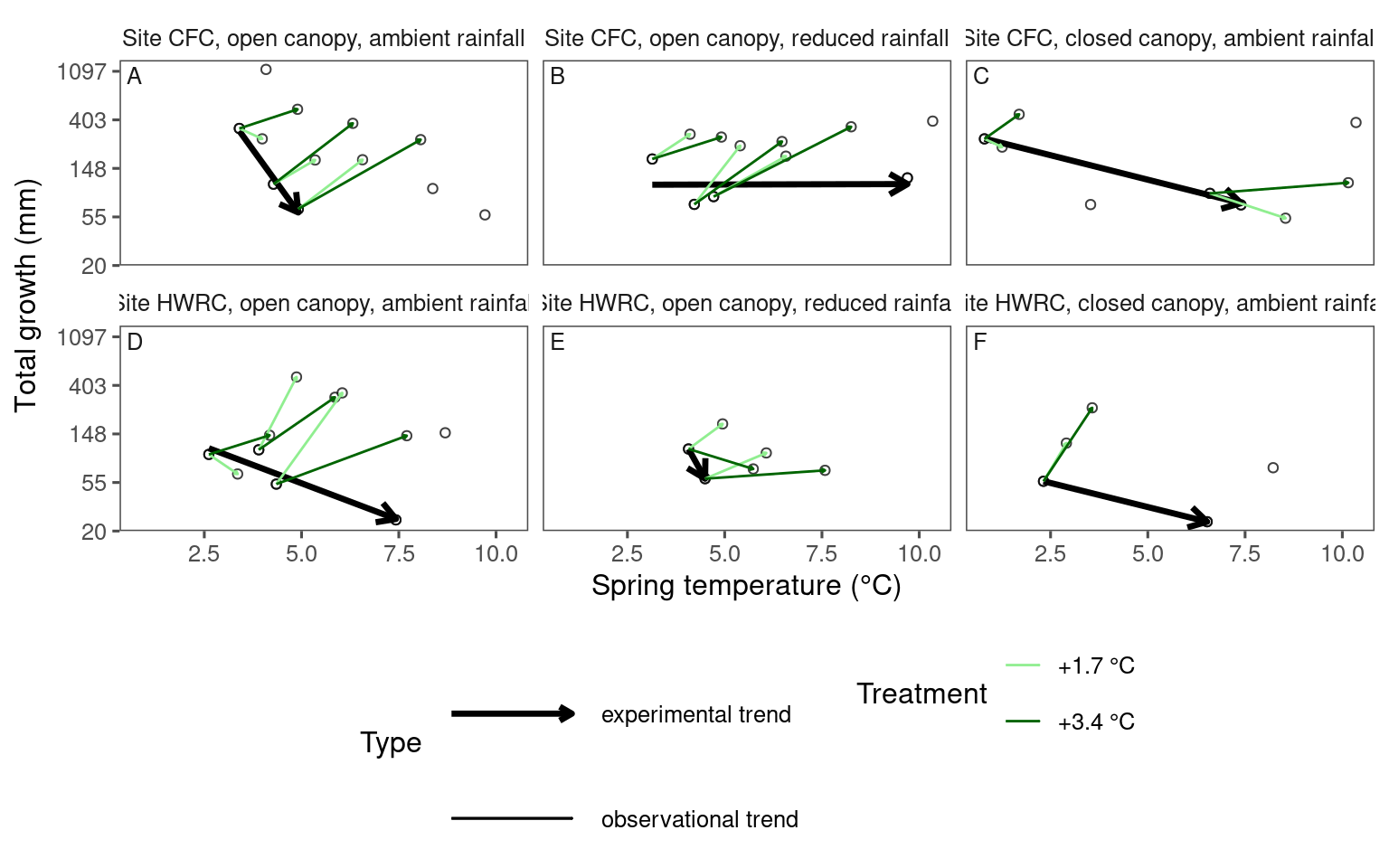
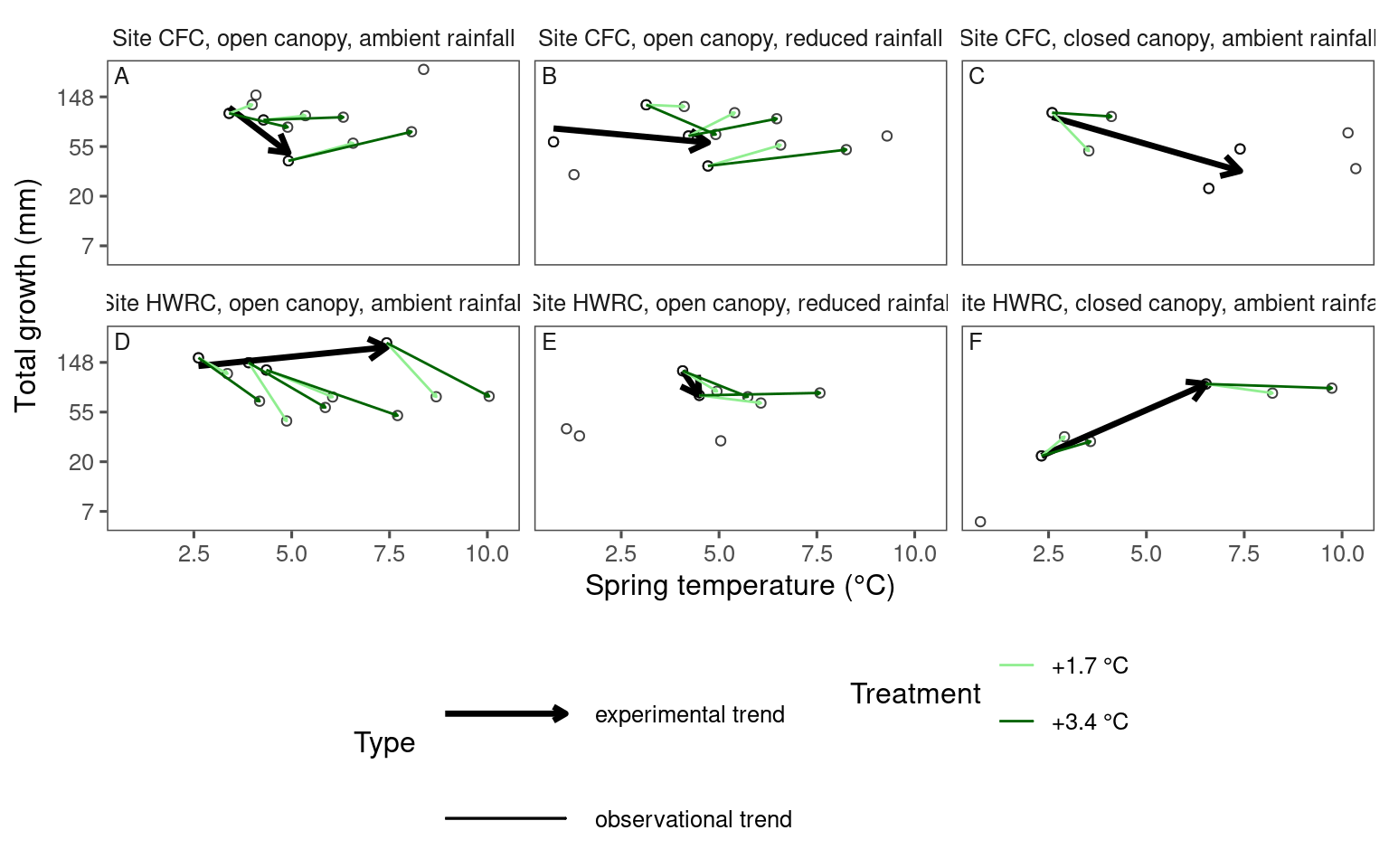
Visualizing experimental and observational trends together with arrows
dat_param_summ <- df_param_all %>%
summ_metrics(level = "site", type = "shoot") # ignore block and plot
dat_climate_spring_summ <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1, level = "site")plot_trend(
df = dat_param_summ %>%
filter(param == "total growth") %>%
filter(species == "acesa"),
dat_climate = dat_climate_spring_summ,
type = "shoot",
side_by_side = F
)
plot_trend(
df = dat_param_summ %>%
filter(param == "total growth") %>%
filter(species == "picgl"),
dat_climate = dat_climate_spring_summ,
type = "shoot",
side_by_side = F
)
Plot comparisons of coefficients
Train model with temperature difference, baseline temperature, and the interaction of the two.
dat_climate_spring_ambient <- calc_climate_ambient(dat_climate = dat_climate_spring, dat_shoot_alive)
dat_climate_spring_diff <- calc_climate_diff(dat_climate = dat_climate_spring, dat_climate_ambient = dat_climate_spring_ambient)
(df_treatment_effect_on_climate <- summ_treatment_effect_on_climate(dat_climate_spring_diff))## # A tibble: 9 × 5
## heat_name water_name canopy temp mois
## <fct> <fct> <fct> <chr> <chr>
## 1 ambient ambient open -0.0391 ± 0.165 0.00346 ± 0.0262
## 2 +1.7 °C ambient open 1.03 ± 0.485 0.00277 ± 0.0253
## 3 +3.4 °C ambient open 1.99 ± 0.815 -0.00772 ± 0.0233
## 4 ambient reduced open 0.000266 ± 0.157 -0.000221 ± 0.0126
## 5 +1.7 °C reduced open 0.837 ± 0.677 0.00332 ± 0.0244
## 6 +3.4 °C reduced open 1.76 ± 0.993 -0.00653 ± 0.0178
## 7 ambient ambient closed -0.00411 ± 0.17 0.00116 ± 0.0384
## 8 +1.7 °C ambient closed 1.07 ± 0.532 0.00251 ± 0.0374
## 9 +3.4 °C ambient closed 2.08 ± 0.97 -0.0033 ± 0.0366Examine number of years
dat_shoot_alive %>%
group_by(species, canopy, water_name) %>%
summarise(n_year = n_distinct(year)) %>%
arrange(n_year)## # A tibble: 45 × 4
## # Groups: species, canopy [31]
## species canopy water_name n_year
## <chr> <fct> <fct> <int>
## 1 fraal open ambient 1
## 2 fraal open reduced 1
## 3 lonmo open ambient 1
## 4 lonmo open reduced 1
## 5 lonta open ambient 1
## 6 lonta open reduced 1
## 7 abiba closed ambient 4
## 8 acene closed ambient 4
## 9 acesa closed ambient 4
## 10 betal closed ambient 4
## # ℹ 35 more rowsdat_shoot_all_cov <- dat_shoot_alive %>%
tidy_shoot_extend() %>%
filter(doy > 90, doy <= 210) %>%
mutate(model = str_c(species, canopy, water_name, sep = "_")) %>%
group_by(species, model) %>%
filter(n_distinct(year) >= 4) %>%
ungroup() %>%
left_join(dat_climate_spring_diff) %>%
drop_na(delta_temp_std, base_temp_std) %>%
group_by(species, model) %>%
mutate(group = str_c(site, year, sep = "_") %>% factor() %>% as.integer()) %>% # site-year level random effects
ungroup() %>%
tidy_treatment_code() %>%
{
# Extract only the `center` and `std` attributes
attributes(.)$center <- attr(dat_climate_spring_diff, "center")
attributes(.)$sd <- attr(dat_climate_spring_diff, "sd")
.
}I used a model with continuous temperature difference (with ambient), continuous baseline temperature, and their interaction.
calc_bayes_all(
data = dat_shoot_all_cov,
independent_priors = F, # do not use species-specific empirical informative priors
uniform_priors = T, # use uniform priors
intui_param = F, # regular parameterization with A, x0, logk
continuous_cov = T, # continuous treatment,
background = T, # background temperature
num_iterations = 50000,
nthin = 5,
path = "alldata/intermediate/shootmodeling/continuous_bg/",
num_cores = 35
)
calc_bayes_derived(path = "alldata/intermediate/shootmodeling/continuous_bg/", num_cores = 35)
plot_bayes_all(path = "alldata/intermediate/shootmodeling/continuous_bg/", num_cores = 35)I also tried a version that experimental treatment was discrete and baseline temperature was continuous.
calc_bayes_all(
data = dat_shoot_all_cov,
independent_priors = F, # do not use species-specific empirical informative priors
uniform_priors = T, # use uniform priors
intui_param = F, # regular parameterization with A, x0, logk
continuous_cov = F, # discrete treatment,
background = T, # background temperature
num_iterations = 50000,
nthin = 5,
path = "alldata/intermediate/shootmodeling/discrete_bg/",
num_cores = 35
)
calc_bayes_derived(path = "alldata/intermediate/shootmodeling/discrete_bg/", num_cores = 35)
plot_bayes_all(path = "alldata/intermediate/shootmodeling/discrete_bg/", num_cores = 35)df_bayes_all <- read_bayes_all(path = "alldata/intermediate/shootmodeling/continuous_bg/", full_factorial = F, derived = T, tidy_mcmc = T) %>%
tidy_model_name() %>%
tidy_species_name()p_bayes_summ <- plot_bayes_summary(
df_bayes_all %>%
filter(response == "total growth") %>%
filter(covariate == "warming"),
background = "full"
)
p_bayes_summ$p_coef_line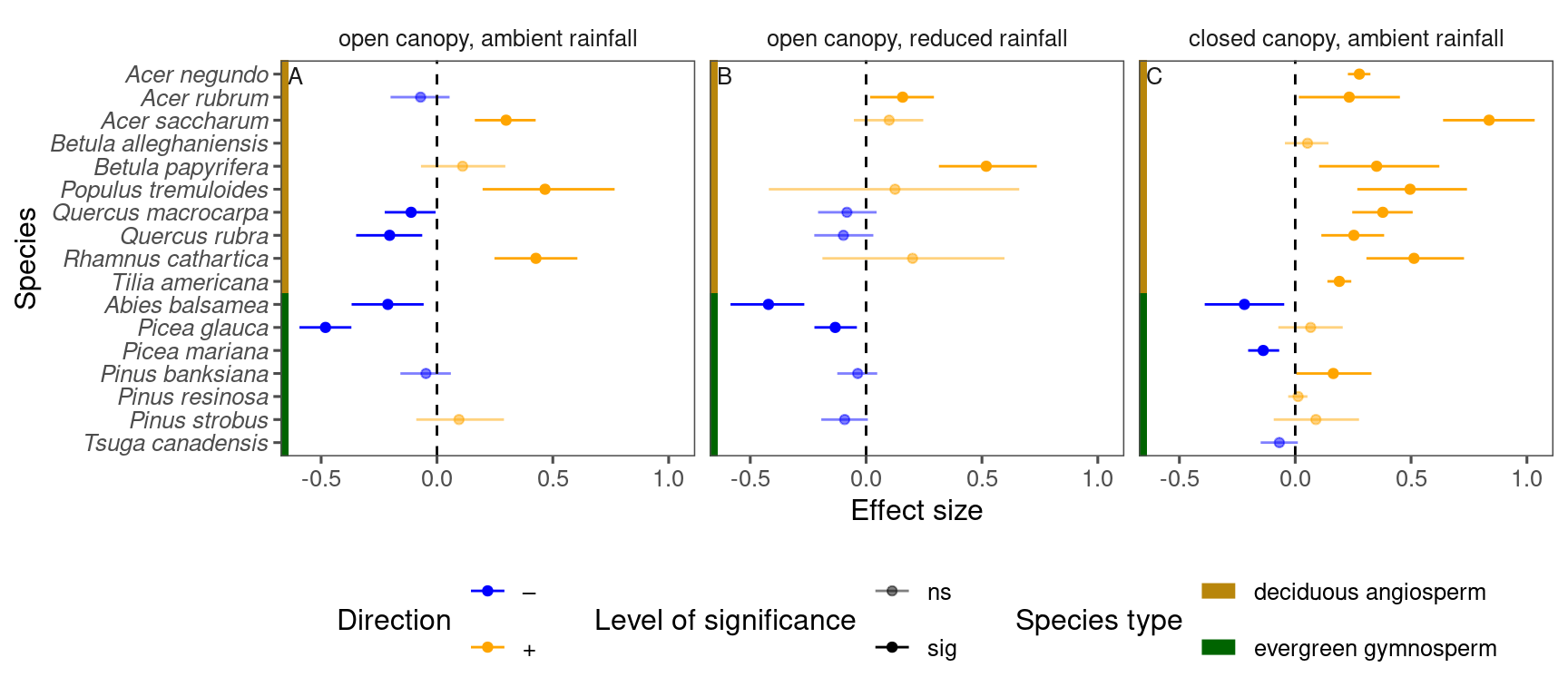
p_bayes_summ <- plot_bayes_summary(
df_bayes_all %>%
filter(response == "total growth") %>%
filter(covariate == "temperature"),
background = "full"
)
p_bayes_summ$p_coef_line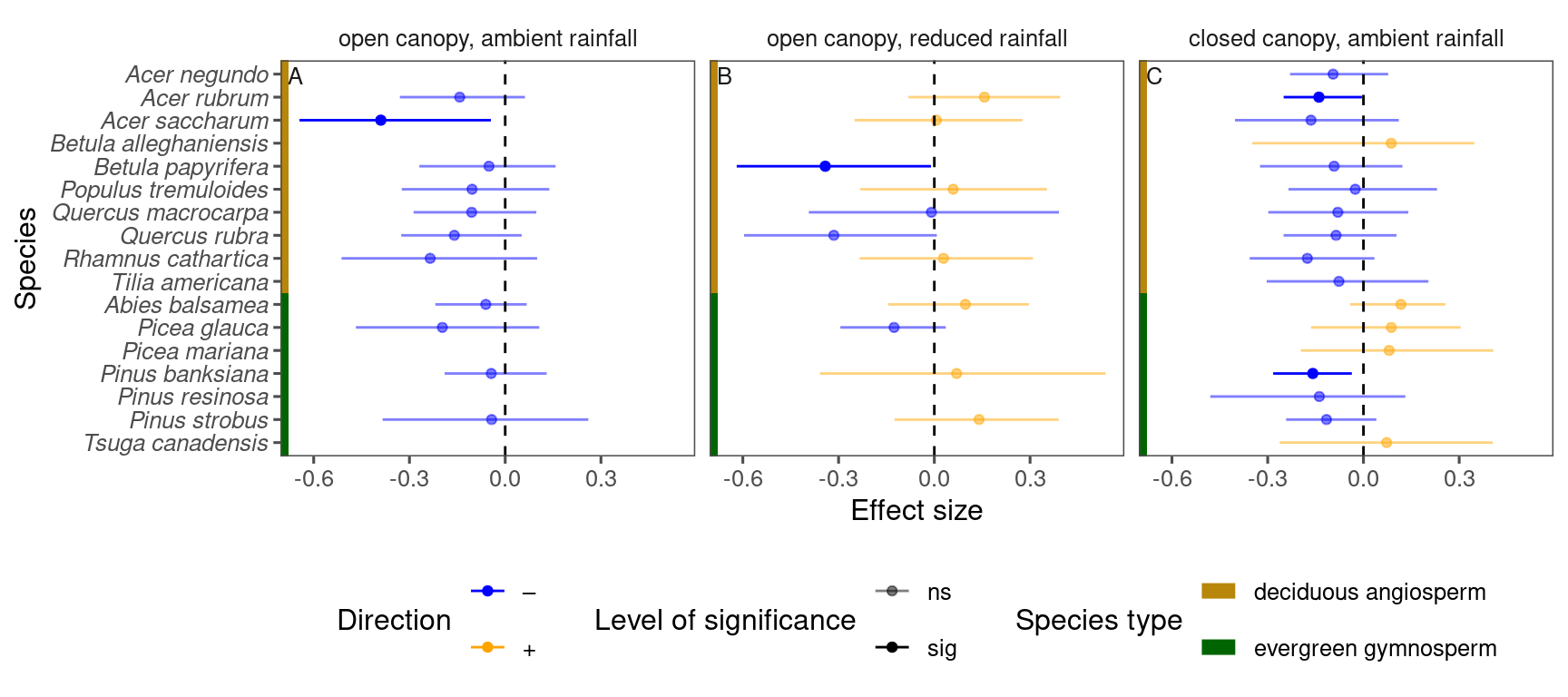
p_bayes_summ <- plot_bayes_summary(
df_bayes_all %>%
filter(response == "start") %>%
filter(covariate == "warming x temperature"),
background = "full"
)
p_bayes_summ$p_coef_line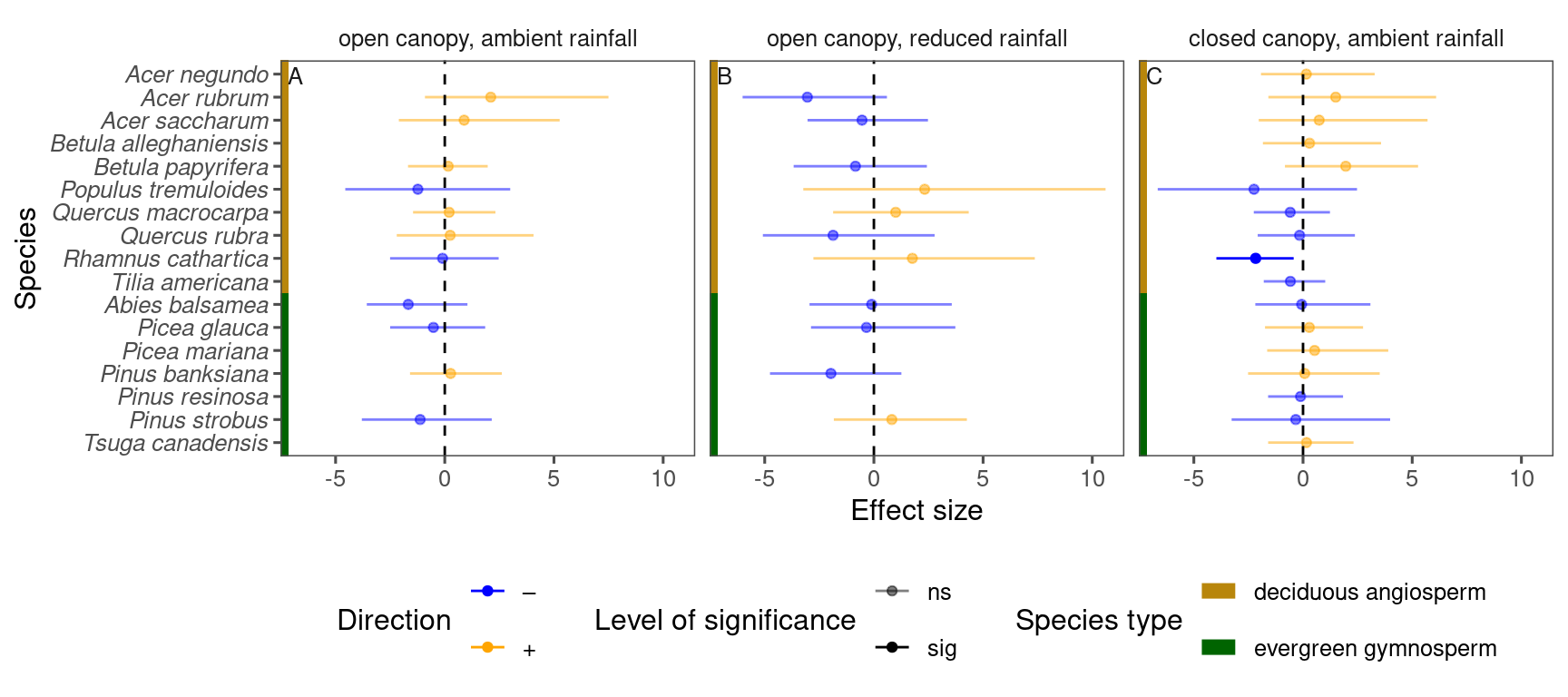
df_compare_trends <- test_compare_trends(df = df_bayes_all, v_response = c("total growth", "midpoint"))plot_trend_compare(df_compare_trends %>% filter(response == "total growth"), type = "shoot", species_label = T, ci = F)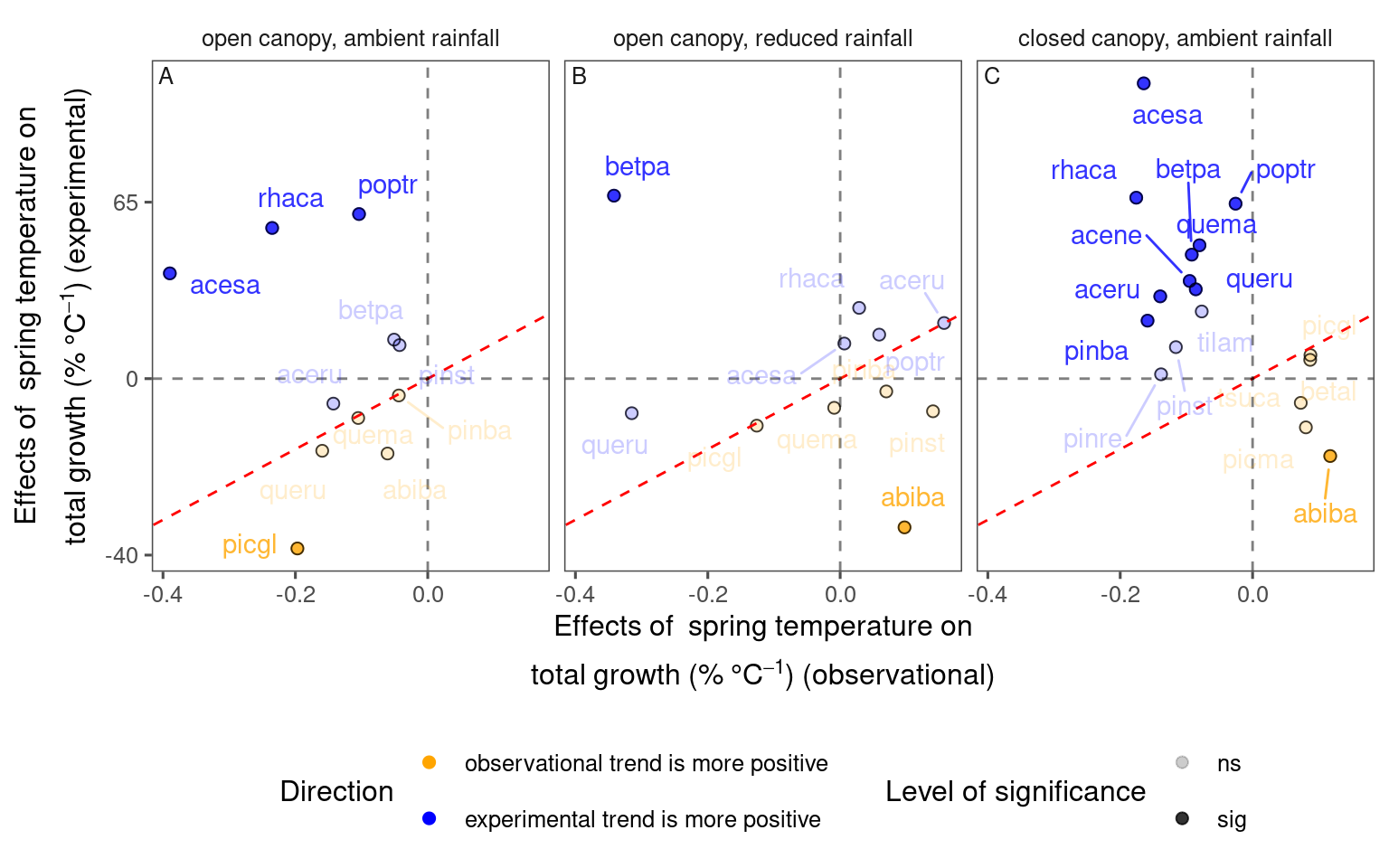
df_compare_trends %>%
filter(response == "total growth") %>%
group_by(sign, sig) %>%
summarise(n = n(), .groups = "drop") %>%
mutate(perc = n / sum(n))## # A tibble: 4 × 4
## sign sig n perc
## <chr> <chr> <int> <dbl>
## 1 + ns 11 0.282
## 2 + sig 13 0.333
## 3 – ns 12 0.308
## 4 – sig 3 0.0769