Background climate
Interannual changes in climatic variables.
dat_climate_annual <- summ_climate_season(dat_climate_daily, date_start = "Jan 1", date_end = "Dec 31", rainfall = 1)
dat_climate_annual_ambient <- calc_climate_ambient(dat_climate_annual, dat_phenophase)
plot_climate_change(dat_climate_annual_ambient, start_year = 2012)
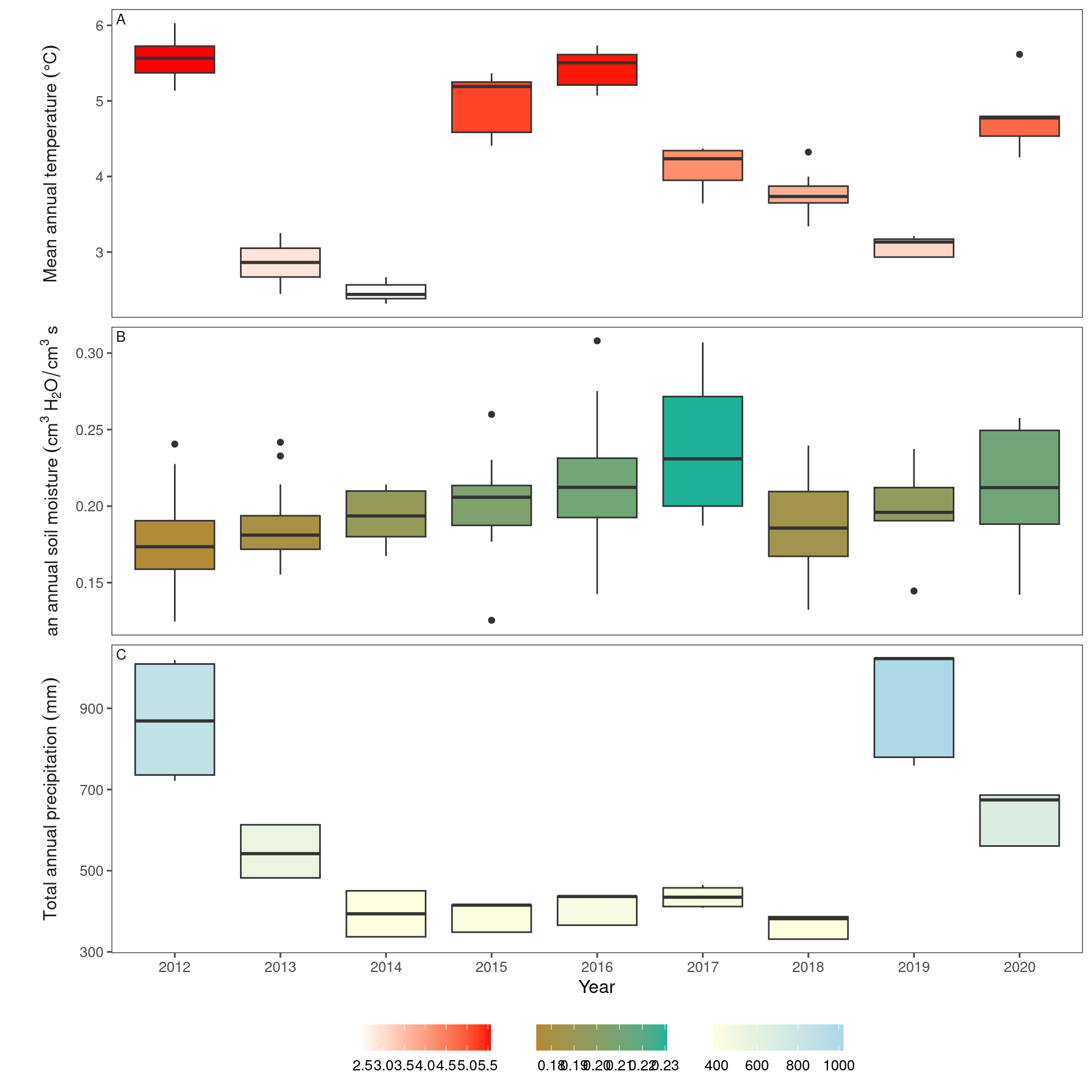
Long-term climate of the two sites.
dat_climate_annual_ambient %>%
group_by(site) %>%
filter(year >= 2012) %>%
summarise(
temp_mean = mean(temp, na.rm = T),
mois_mean = mean(mois, na.rm = T),
prcp_mean = mean(prcp, na.rm = T),
temp_std = sd(temp, na.rm = T),
mois_std = sd(mois, na.rm = T),
prcp_std = sd(prcp, na.rm = T)
)
## # A tibble: 2 × 7
## site temp_mean mois_mean prcp_mean temp_std mois_std prcp_std
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 cfc 4.36 0.196 595. 1.10 0.0214 239.
## 2 hwrc 4.10 0.201 471. 1.16 0.0509 158.
Factorial design
There are still problems with the block and plot IDs.
dat_shoot %>%
pull(plot) %>%
unique() %>%
length()
## [1] 72
dat_shoot %>%
group_by(site, canopy, heat_name, water_name) %>%
summarise(
n_block = n_distinct(block),
n_plot = n_distinct(plot)
)
## # A tibble: 18 × 6
## # Groups: site, canopy, heat_name [12]
## site canopy heat_name water_name n_block n_plot
## <chr> <chr> <fct> <fct> <int> <int>
## 1 cfc closed ambient ambient 3 6
## 2 cfc closed +1.7 °C ambient 3 6
## 3 cfc closed +3.4 °C ambient 3 6
## 4 cfc open ambient ambient 3 3
## 5 cfc open ambient reduced 3 3
## 6 cfc open +1.7 °C ambient 3 3
## 7 cfc open +1.7 °C reduced 3 3
## 8 cfc open +3.4 °C ambient 3 3
## 9 cfc open +3.4 °C reduced 3 3
## 10 hwrc closed ambient ambient 3 6
## 11 hwrc closed +1.7 °C ambient 3 6
## 12 hwrc closed +3.4 °C ambient 3 6
## 13 hwrc open ambient ambient 3 3
## 14 hwrc open ambient reduced 3 3
## 15 hwrc open +1.7 °C ambient 3 4
## 16 hwrc open +1.7 °C reduced 3 3
## 17 hwrc open +3.4 °C ambient 3 4
## 18 hwrc open +3.4 °C reduced 3 3
dat_shoot %>%
distinct(site, canopy, heat_name, water_name, block, plot) %>%
arrange(plot)
## # A tibble: 74 × 6
## site canopy heat_name water_name block plot
## <chr> <chr> <fct> <fct> <chr> <chr>
## 1 cfc closed +3.4 °C ambient a a1
## 2 cfc closed +1.7 °C ambient a a2
## 3 cfc closed ambient ambient a a3
## 4 cfc closed ambient ambient a a5
## 5 cfc closed +1.7 °C ambient a a6
## 6 cfc closed +3.4 °C ambient a a7
## 7 cfc closed +1.7 °C ambient b b2
## 8 cfc closed ambient ambient b b3
## 9 cfc closed +3.4 °C ambient b b4
## 10 cfc closed +3.4 °C ambient b b5
## # ℹ 64 more rows
Seedling planting
dat_phenophase %>%
group_by(cohort) %>%
summarise(n_seedling = n_distinct(barcode)) %>%
arrange(cohort)
## # A tibble: 7 × 2
## cohort n_seedling
## <dbl> <int>
## 1 2008 2487
## 2 2011 63
## 3 2012 1175
## 4 2013 302
## 5 2014 1791
## 6 2017 1483
## 7 2020 23
dat_phenophase %>%
mutate(age = year - cohort) %>%
group_by(barcode) %>%
summarise(max_age = max(age)) %>%
pull(max_age) %>%
summary()
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.00 2.00 3.00 3.07 4.00 11.00
Goodness-of-fit
dat_shoot_extend <- dat_shoot %>% tidy_shoot_extend()
dat_all <- dat_shoot_extend %>%
filter(doy > 90, doy <= 210) %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
group_by(species) %>%
mutate(group = str_c(site, year, sep = "_") %>% factor() %>% as.integer()) %>% # site-year level random effects
ungroup() %>%
tidy_treatment_code()
df_bayes_pred_all <- read_bayes_all(path = "alldata/intermediate/shootmodeling/uni/", full_factorial = T, content = "predict")
# Calculate spearman R^2
(df_R2_species <- inner_join(
dat_all %>% select(species, site, year, heat_trt, water_trt, canopy_code, doy, obs = shoot),
df_bayes_pred_all %>% select(species, site, year, heat_trt, water_trt, canopy_code, doy, pred = pred_median)
) %>%
group_by(species) %>%
summarise(
R2 = cor(obs, pred, method = "spearman")^2,
n = n()
) %>%
arrange(desc(R2)))
## # A tibble: 11 × 3
## species R2 n
## <chr> <dbl> <int>
## 1 pinba 0.773 8057
## 2 picgl 0.743 10411
## 3 quema 0.720 10366
## 4 pinst 0.706 10288
## 5 betpa 0.698 10433
## 6 abiba 0.691 9974
## 7 aceru 0.668 10352
## 8 rhaca 0.631 7221
## 9 queru 0.623 10296
## 10 poptr 0.593 6715
## 11 acesa 0.585 10418
(df_R2_overall <- inner_join(
dat_all %>% select(species, site, year, heat_trt, water_trt, canopy_code, doy, obs = shoot),
df_bayes_pred_all %>% select(species, site, year, heat_trt, water_trt, canopy_code, doy, pred = pred_median)
) %>%
summarise(
R2 = cor(obs, pred, method = "spearman")^2,
n = n()
))
## # A tibble: 1 × 2
## R2 n
## <dbl> <int>
## 1 0.709 104531
Report coefficients
df_bayes_all <- read_bayes_all(path = "alldata/intermediate/shootmodeling/uni/", full_factorial = T, derived = T, tidy_mcmc = T) %>%
tidy_species_name()
df_coef_summ <- summ_mcmc(df_bayes_all)
summ_coef(df_coef_summ, response = "midpoint", covariate = "warming")
## # A tibble: 11 × 10
## param species response covariate median lower upper sig sign category
## <chr> <fct> <fct> <fct> <dbl> <dbl> <dbl> <chr> <chr> <fct>
## 1 beta_xmi… queru midpoint warming -3.64 -4.56 -2.80 sig – negativ…
## 2 beta_xmi… acesa midpoint warming -3.25 -4.57 -2.08 sig – negativ…
## 3 beta_xmi… quema midpoint warming -3.10 -3.82 -2.25 sig – negativ…
## 4 beta_xmi… picgl midpoint warming -3.08 -4.02 -2.17 sig – negativ…
## 5 beta_xmi… pinst midpoint warming -3.06 -4.51 -1.82 sig – negativ…
## 6 beta_xmi… abiba midpoint warming -3.02 -4.40 -1.75 sig – negativ…
## 7 beta_xmi… betpa midpoint warming -2.79 -4.31 -1.35 sig – negativ…
## 8 beta_xmi… rhaca midpoint warming -2.65 -4.53 -0.919 sig – negativ…
## 9 beta_xmi… pinba midpoint warming -2.57 -3.57 -1.54 sig – negativ…
## 10 beta_xmi… aceru midpoint warming -2.56 -3.65 -1.40 sig – negativ…
## 11 beta_xmi… poptr midpoint warming -2.47 -5.00 0.444 ns – negativ…
# omitting other summarise here
Effects on timing
summ_coef(df_coef_summ, response = "midpoint", covariate = "warming")
summ_coef(df_coef_summ, response = "midpoint", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "start", covariate = "warming")
summ_coef(df_coef_summ, response = "start", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "end", covariate = "warming")
summ_coef(df_coef_summ, response = "end", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "midpoint", covariate = "closed")
summ_coef(df_coef_summ, response = "midpoint", covariate = "drying")
summ_coef(df_coef_summ, response = "midpoint", covariate = "warming x drying")
Effects on pace
summ_coef(df_coef_summ, response = "rate", covariate = "warming")
summ_coef(df_coef_summ, response = "rate", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "duration", covariate = "warming")
summ_coef(df_coef_summ, response = "duration", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "speed", covariate = "warming")
summ_coef(df_coef_summ, response = "speed", covariate = "warminasymptoteg | closed")
summ_coef(df_coef_summ, response = "rate", covariate = "closed")
summ_coef(df_coef_summ, response = "duration", covariate = "closed")
summ_coef(df_coef_summ, response = "speed", covariate = "closed")
Effect on magnitude
summ_coef(df_coef_summ, response = "asymptote", covariate = "warming")
summ_coef(df_coef_summ, response = "asymptote", covariate = "warming | closed")
summ_coef(df_coef_summ, response = "asymptote", covariate = "closed")
summ_coef(df_coef_summ, response = "asymptote", covariate = "drying")