Re-examine Montgomery et al. (2020)
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1)
dat_climate_spring_ambient <- calc_climate_ambient(dat_climate = dat_climate_spring, dat_phenophase)
write_rds(dat_climate_spring_ambient, "alldata/intermediate/climate_spring_ambient.rds")
dat_climate_spring_ambient <- read_rds("alldata/intermediate/climate_spring_ambient.rds")
dat_phenophase_time_summ <- summ_phenophase_time(dat_phenophase_time_clean)
dat_phenophase_diff_summ <- calc_phenophase_diff(dat_phenophase_time_summ)
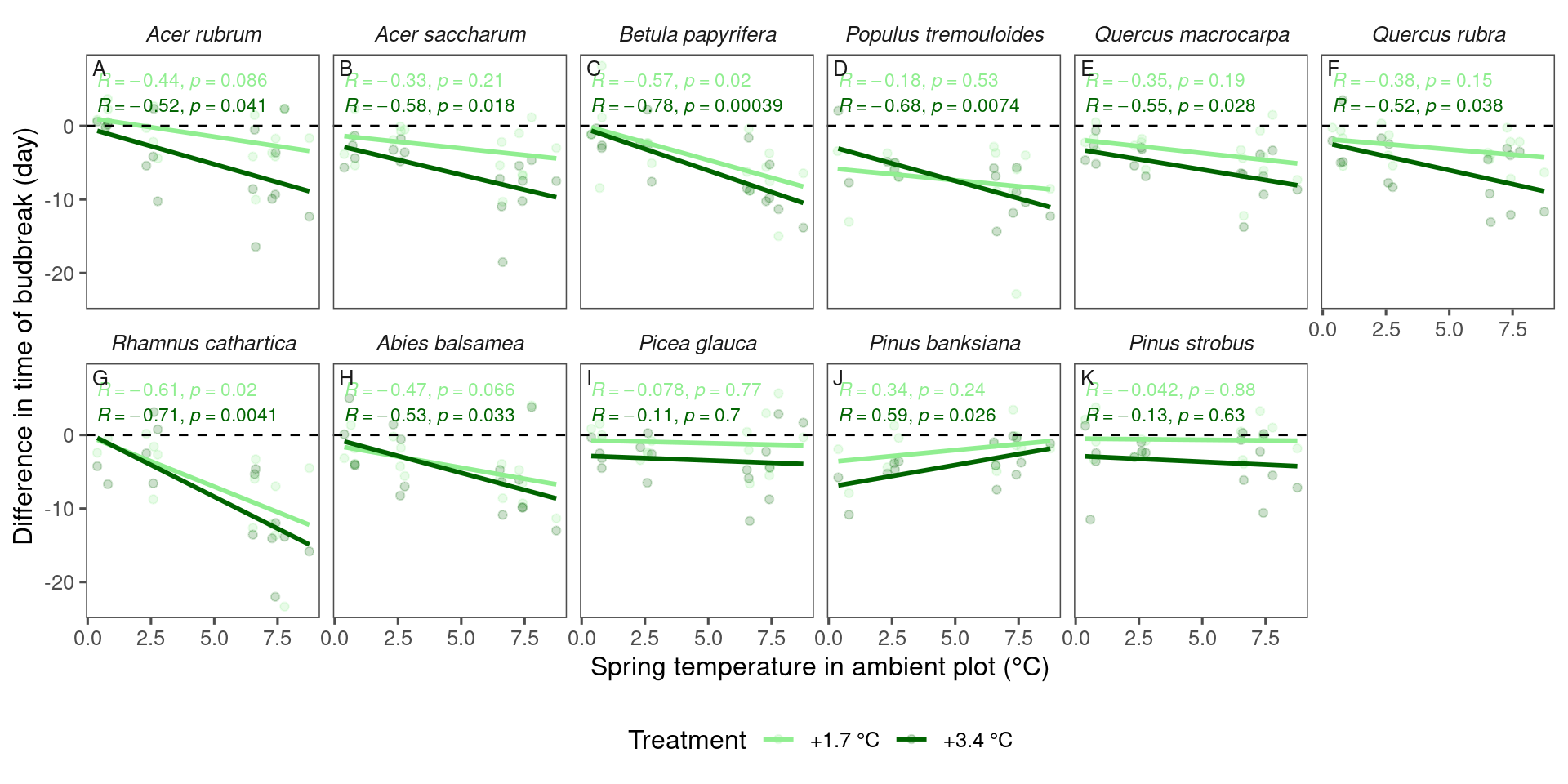
dat_phenophase_diff_summ %>%
filter(phenophase == "budbreak") %>%
filter(year %in% 2009:2013) %>%
tidy_species_name() %>%
tidy_phenophase_name() %>%
plot_diff_background(dat_climate_spring_ambient, var_x = "temp", pool_sp = F)
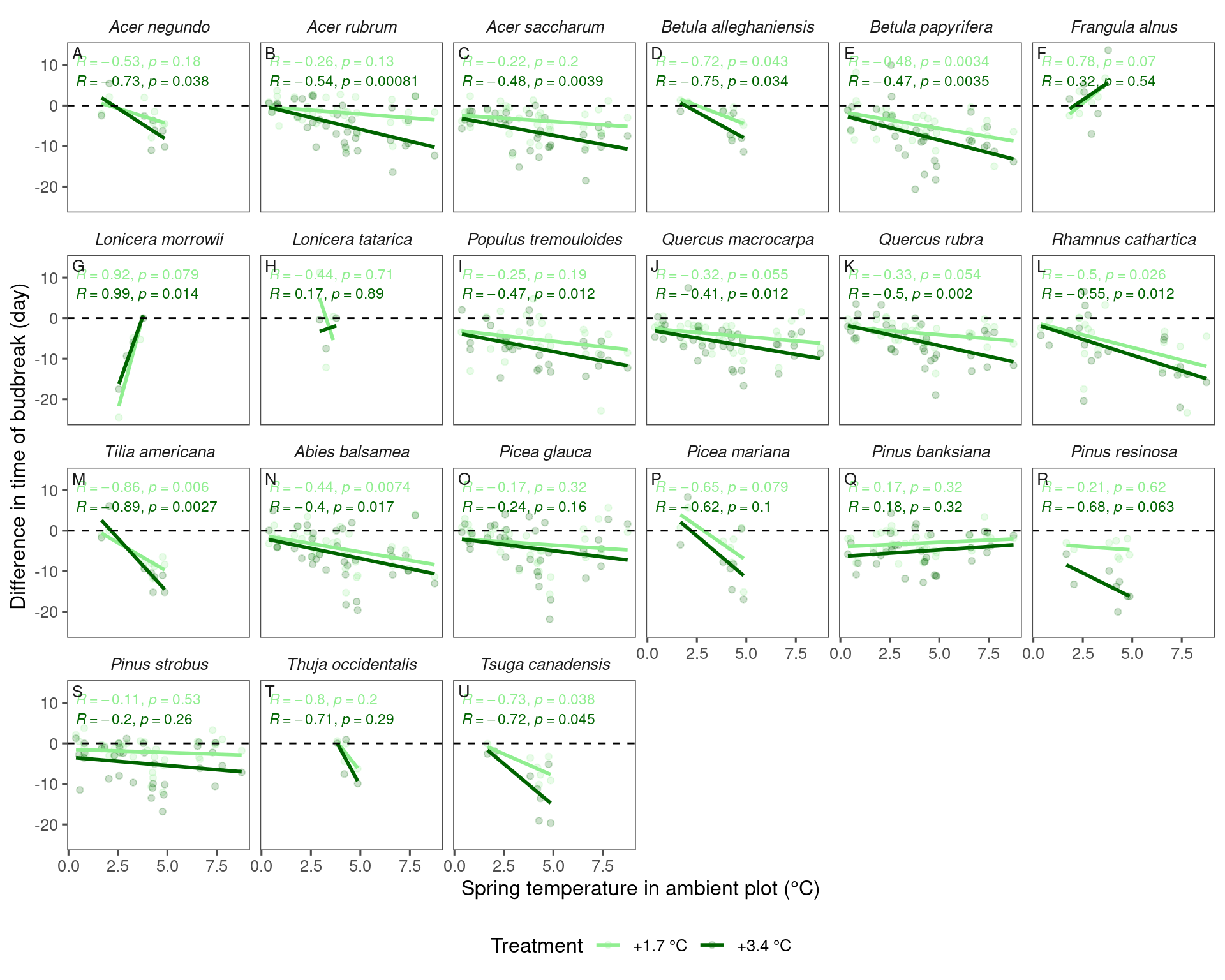
dat_phenophase_diff_summ %>%
filter(phenophase == "budbreak") %>%
tidy_species_name() %>%
tidy_phenophase_name() %>%
plot_diff_background(dat_climate_spring_ambient, var_x = "temp", pool_sp = F)
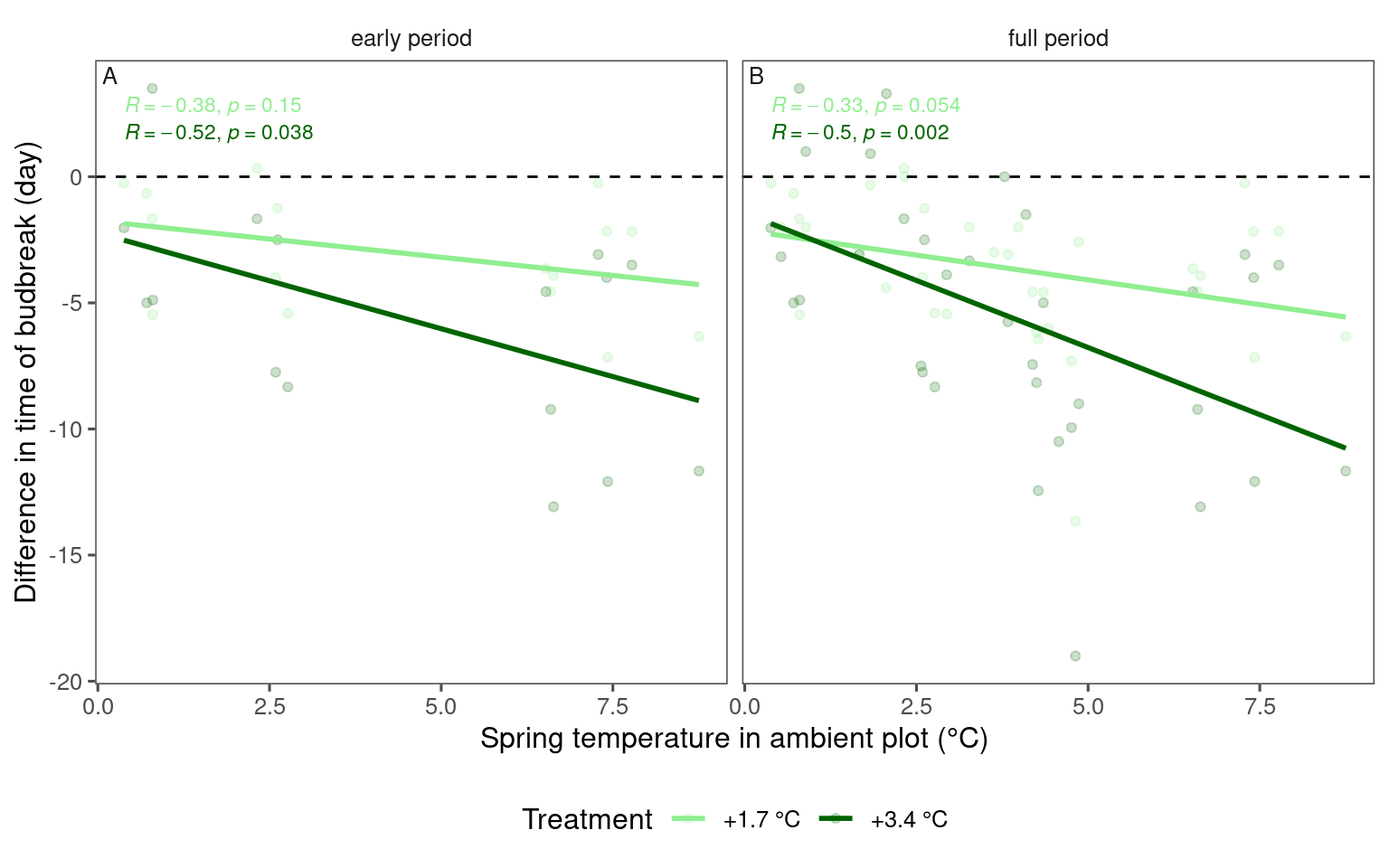
bind_rows(
dat_phenophase_diff_summ %>%
filter(phenophase == "budbreak") %>%
filter(year %in% 2009:2013) %>%
filter(species == "queru") %>%
mutate(period = "early period"),
dat_phenophase_diff_summ %>%
filter(phenophase == "budbreak") %>%
filter(species == "queru") %>%
mutate(period = "full period")
) %>%
tidy_species_name() %>%
tidy_phenophase_name() %>%
plot_diff_background(dat_climate_spring_ambient, var_x = "temp", pool_sp = F)
Spring model
\begin{aligned} y_{i,t,s} &\sim \mathcal{N}(\mu_{i,t,s}, \sigma^2) \newline \mu_{i,t,s} &= \mu + \beta_1 W_i + \beta_2 D_i + \beta_3 (W_i \times D_i) + \beta_4 C_i + \beta_5 (W_i \times C_i) + \newline & \quad \beta_6 T_i + \beta_7 (W_i \times T_i) + \beta_8 (T_i \times C_i) + \beta_9 \theta_i + \alpha_{t,s} \newline \alpha_{t,s} &\sim \mathcal{N}(0, \tau^2) \newline \newline \mu &\sim \mathcal{U}(1, 180) \newline \beta_{1:5} &\sim \mathcal{N}(0, 10) \newline \sigma &\sim \mathcal{N}(0, 4) \newline \tau &\sim \mathcal{N}(0, 4) \end{aligned}
dat_phenophase_time_cov <- dat_phenophase_time_clean %>%
tidy_treatment_code() %>%
left_join(dat_climate_spring_ambient, by = c("site", "canopy", "block", "year")) %>%
group_by(species) %>%
mutate(group = site %>% factor() %>% as.integer()) %>% # use site as random effects, not site-year
ungroup()
test_phenophase_lme(data = dat_phenophase_time_cov, path = "alldata/intermediate/phenophase/bg/", season = "spring", chains = 1, iter = 4000, version = 3, num_cores = 5) # v3 with interactions
tidy_stanfit_all(season = "spring", path = "alldata/intermediate/phenophase/bg/")
calc_bayes_derived(path = "alldata/intermediate/phenophase/bg/", season = "spring", type = "phenophase", random = F, num_cores = 20)
df_lme_all <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "spring", full_factorial = T, derived = F, tidy_mcmc = F) %>%
tidy_phenophase_name(season = "spring")
p_lme_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_lme_all %>% filter(species == "queru"), plot_corr = F)
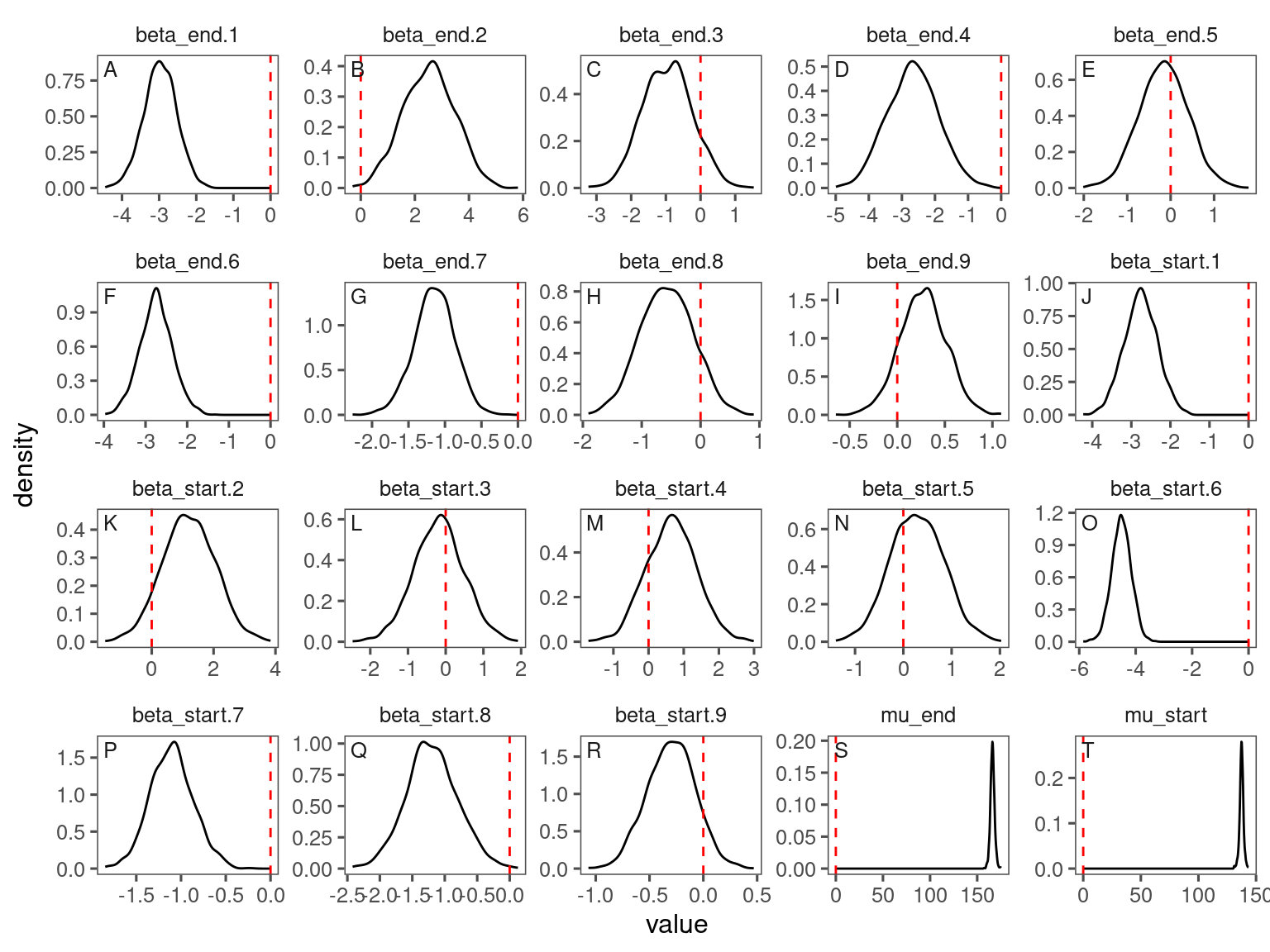
p_lme_diagnostics$p_MCMC
p_lme_diagnostics$p_posterior
df_lme_all_derived <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "spring", full_factorial = T, derived = T, tidy_mcmc = T) %>%
tidy_species_name() %>%
tidy_phenophase_name(season = "spring")
p_lme_summ <- plot_bayes_summary(
df_lme_all_derived,
background = "only",
plot_corr = F
)
p_lme_summ$p_coef_line
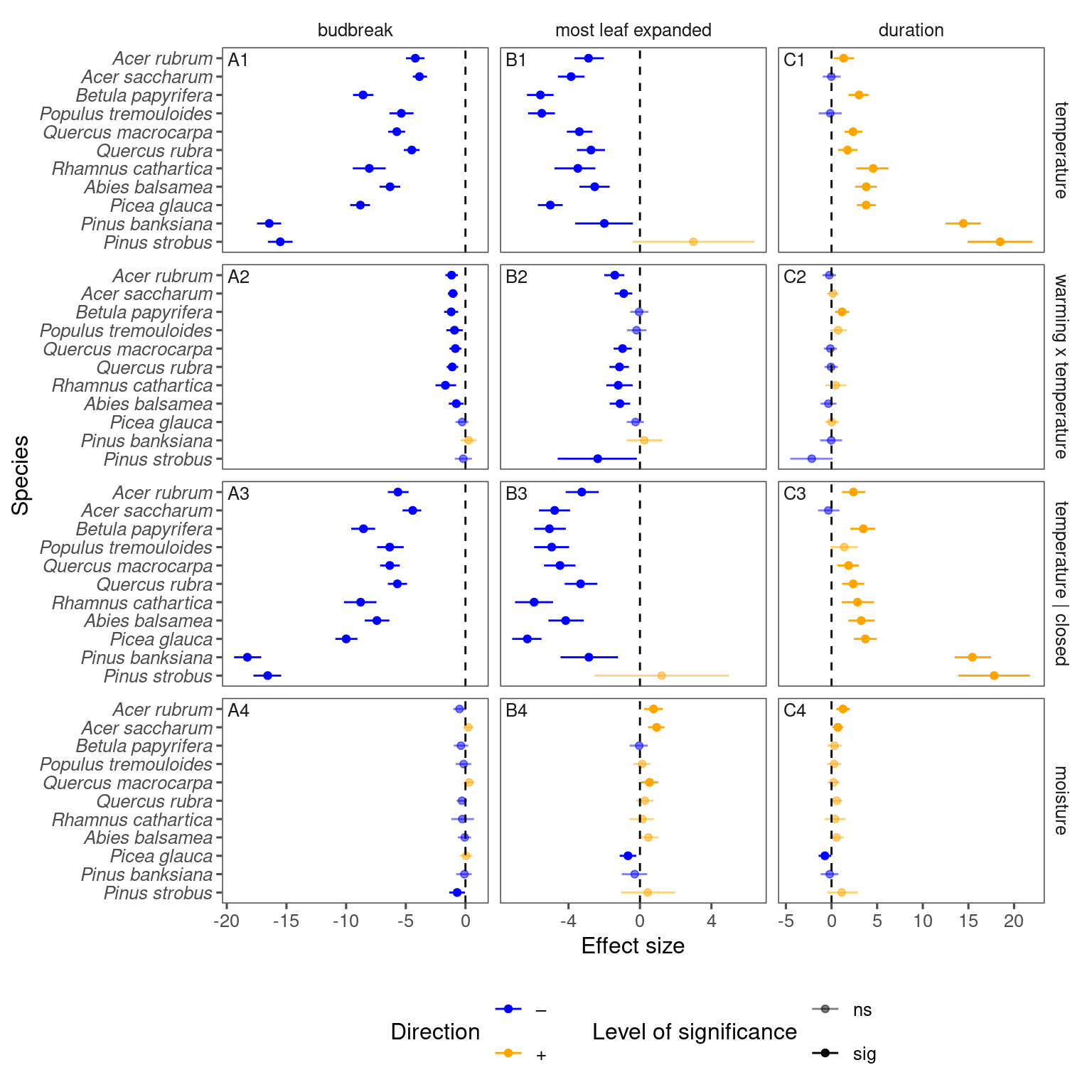
p_lme_summ <- plot_bayes_summary(
df_lme_all_derived,
background = "no",
plot_corr = F
)
p_lme_summ$p_coef_line
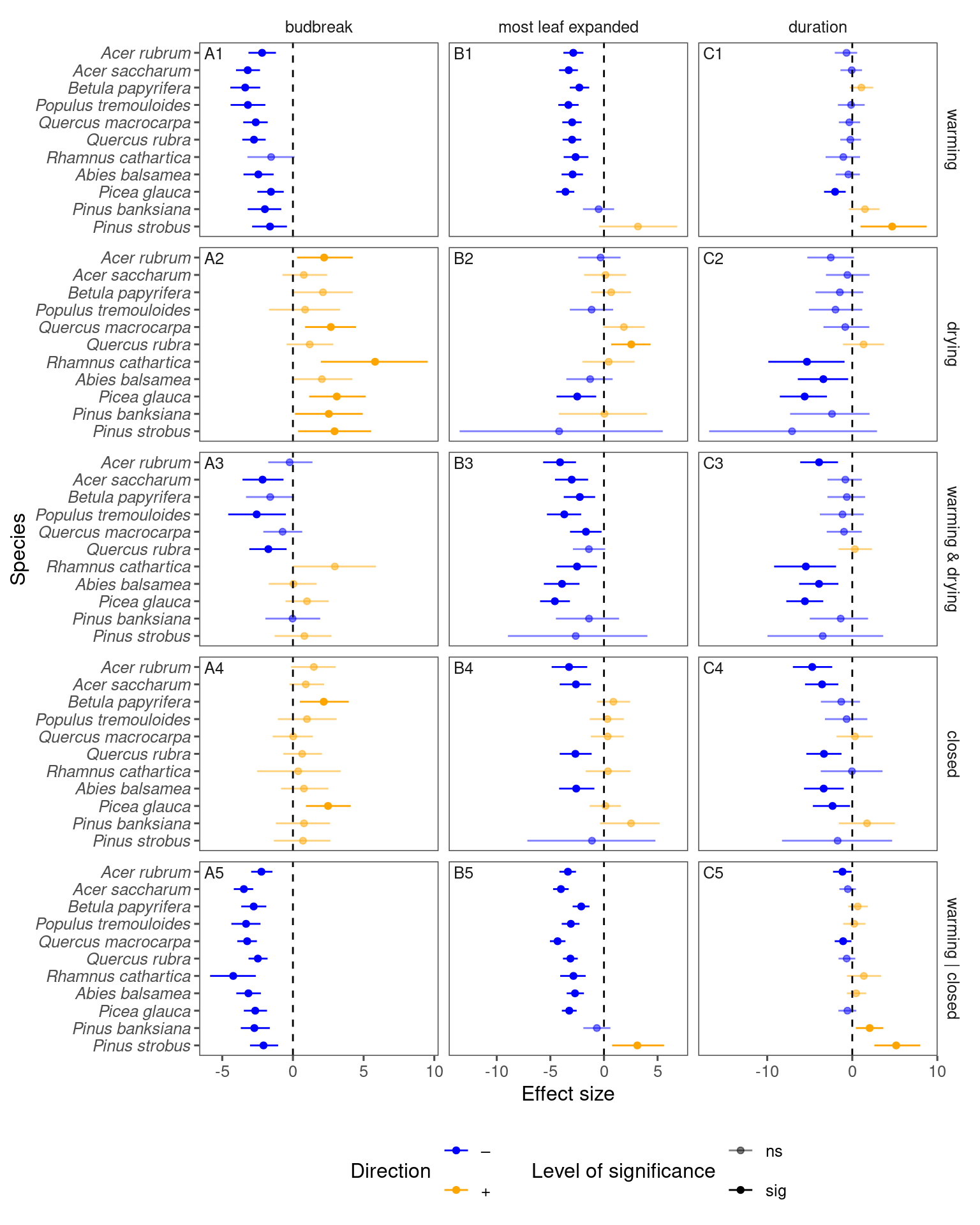
Fall model
\begin{aligned} y_{i,t,s} &\sim \mathcal{N}(\mu_{i,t,s}, \sigma^2) \newline \mu_{i,t,s} &= \mu + \beta_1 W_i + \beta_2 D_i + \beta_3 (W_i \times D_i) + \beta_4 C_i + \beta_5 (W_i \times C_i) + \newline & \quad \beta_6 T_i + \beta_7 (W_i \times T_i) + \beta_8 (T_i \times C_i) + \beta_9 \theta_i + \alpha_{t,s} \newline \alpha_{t,s} &\sim \mathcal{N}(0, \tau^2) \newline \newline \mu &\sim \mathcal{U}(181, 365) \newline \beta_{1:5} &\sim \mathcal{N}(0, 10) \newline \sigma &\sim \mathcal{N}(0, 4) \newline \tau &\sim \mathcal{N}(0, 4) \end{aligned}
dat_climate_fall <- summ_climate_season(dat_climate_daily, date_start = "Jun 15", date_end = "Sep 15", rainfall = 0)
dat_climate_fall_ambient <- calc_climate_ambient(dat_climate = dat_climate_fall, dat_phenophase)
write_rds(dat_climate_fall_ambient, "alldata/intermediate/climate_fall_ambient.rds")
dat_phenophase_time_cov <- dat_phenophase_time_clean %>%
tidy_treatment_code() %>%
left_join(dat_climate_fall_ambient, by = c("site", "canopy", "block", "year")) %>%
group_by(species) %>%
mutate(group = site %>% factor() %>% as.integer()) %>% # use site as random effects, not site-year
ungroup()
test_phenophase_lme(data = dat_phenophase_time_cov, path = "alldata/intermediate/phenophase/bg/", season = "fall", chains = 1, iter = 4000, version = 3, num_cores = 5) # v3 with interactions
tidy_stanfit_all(season = "fall", path = "alldata/intermediate/phenophase/bg/")
calc_bayes_derived(path = "alldata/intermediate/phenophase/bg/", season = "fall", type = "phenophase", random = F, num_cores = 20)
df_lme_all <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "fall", full_factorial = T, derived = F, tidy_mcmc = F) %>%
tidy_phenophase_name(season = "fall")
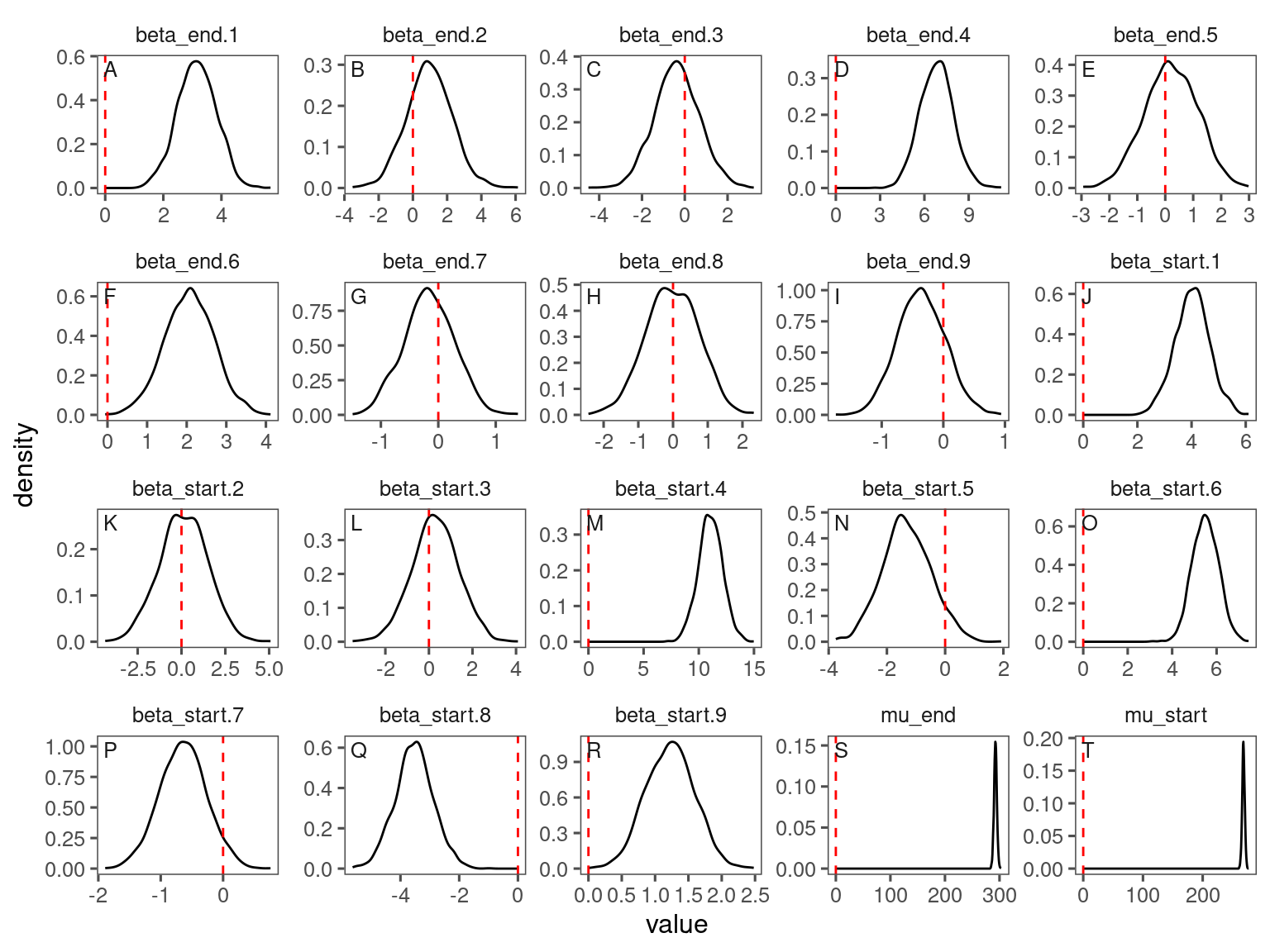
p_lme_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_lme_all %>% filter(species == "queru"), plot_corr = F)
p_lme_diagnostics$p_MCMC
p_lme_diagnostics$p_posterior
df_lme_all_derived <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "fall", full_factorial = T, derived = T, tidy_mcmc = T) %>%
tidy_species_name() %>%
tidy_phenophase_name(season = "fall")
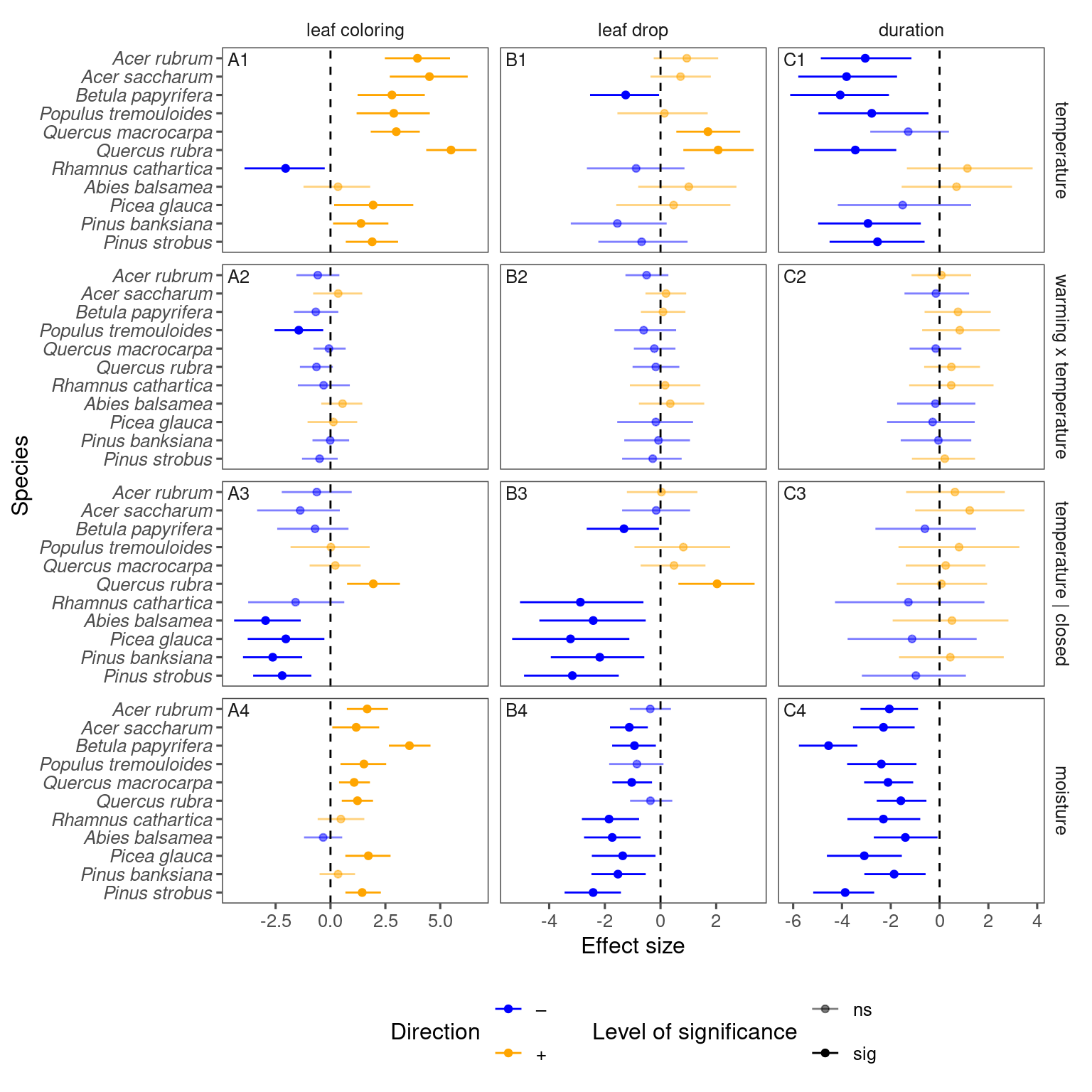
p_lme_summ <- plot_bayes_summary(
df_lme_all_derived,
background = "only",
plot_corr = F
)
p_lme_summ$p_coef_line
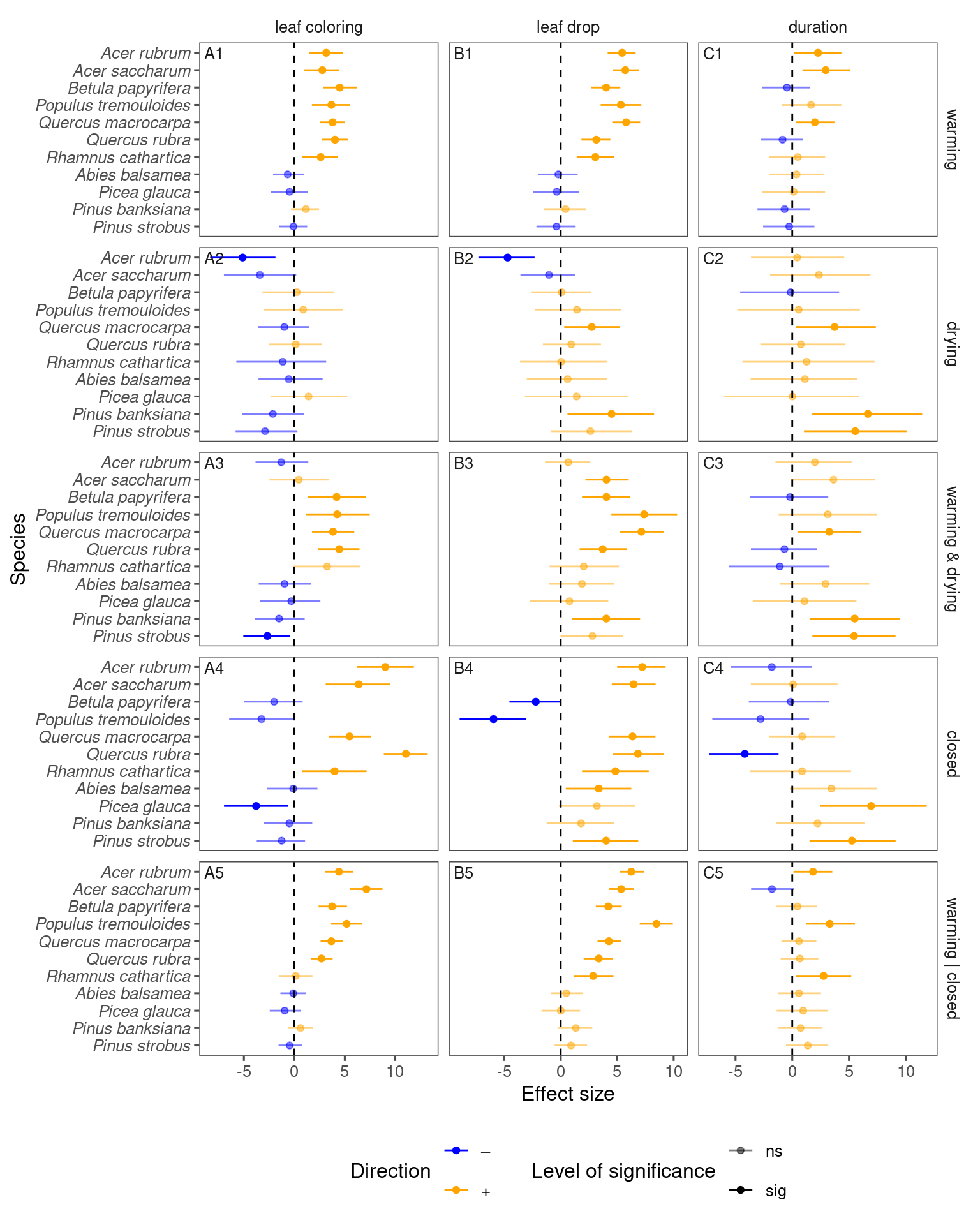
p_lme_summ <- plot_bayes_summary(
df_lme_all_derived,
background = "no",
plot_corr = F
)
p_lme_summ$p_coef_line
Winter vs. spring warming
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1, group_vars = c("site", "canopy", "heat", "water", "block", "plot", "year"))
dat_climate_winter <- summ_climate_season(dat_climate_daily, date_start = "Dec 15", start_rollback = 1, date_end = "Feb 15", rainfall = 1, group_vars = c("site", "canopy", "heat", "water", "block", "plot", "year"))
dat_phenophase_time_clean %>%
filter(phenophase == "budbreak") %>%
tidy_phenophase_name() %>%
tidy_species_name() %>%
plot_winter_spring(dat_climate_winter, dat_climate_spring)
