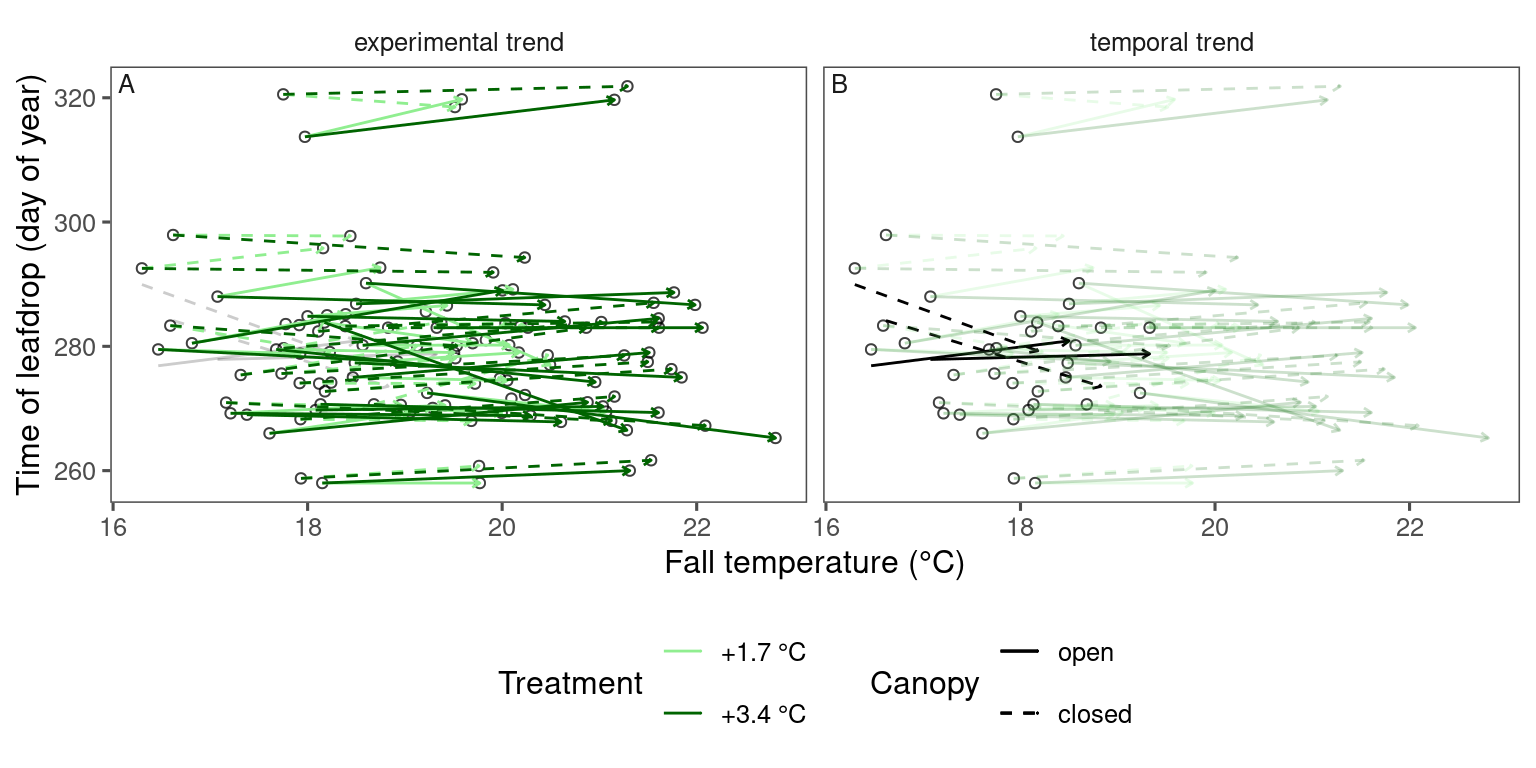
Plot arrows
dat_phenophase_time_summ <- dat_phenophase_time_clean %>%
summ_phenophase_time(group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "species", "phenophase", "year")) # ignore block and plot
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))
dat_climate_fall <- summ_climate_season(dat_climate_daily, date_start = "Jun 15", date_end = "Sep 15", rainfall = 0, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))
plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "budbreak") %>%
filter(species == "pinba"),
dat_climate = dat_climate_spring
)
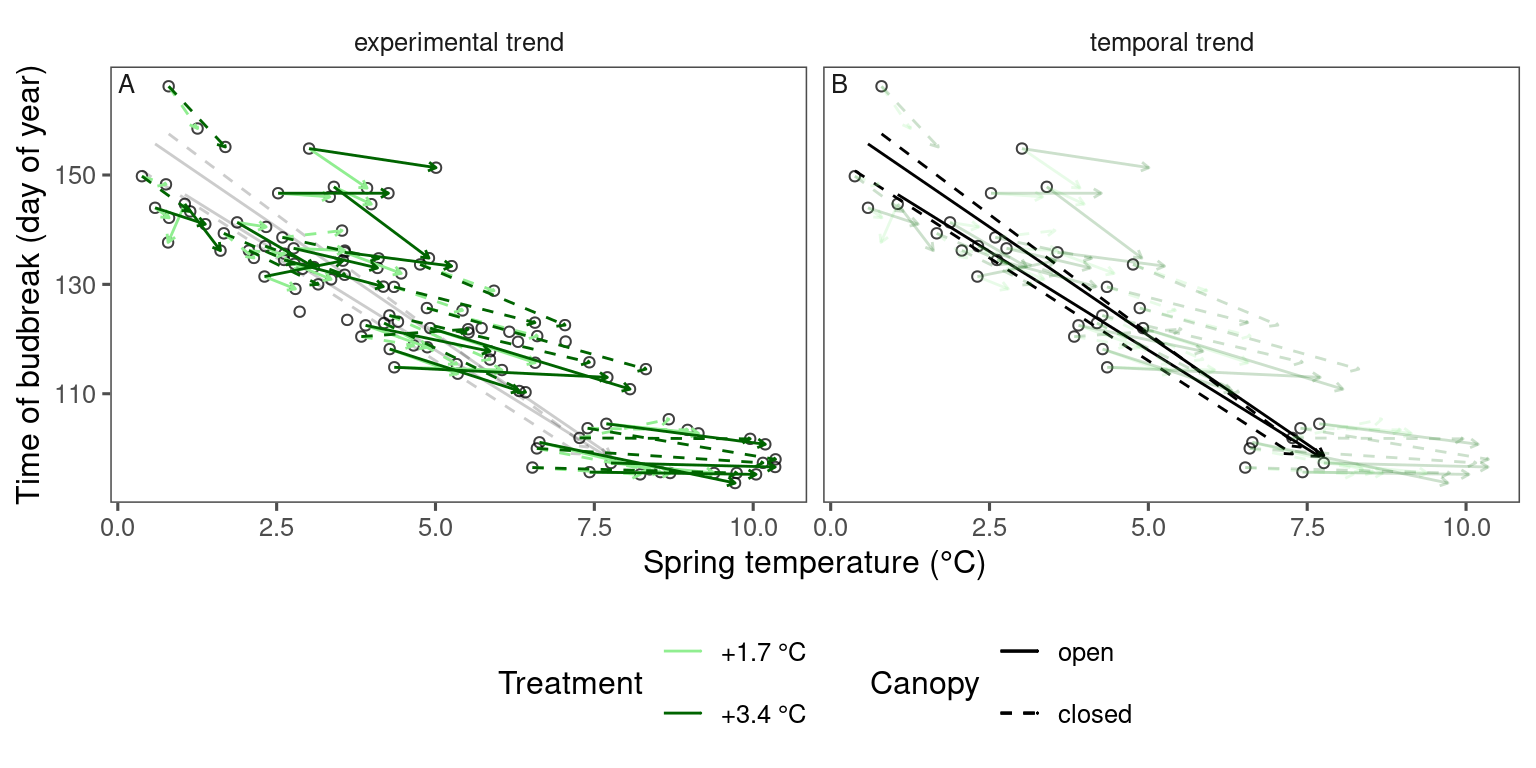
plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "mostleaf") %>%
filter(species == "poptr"),
dat_climate = dat_climate_spring,
var = "heat"
)
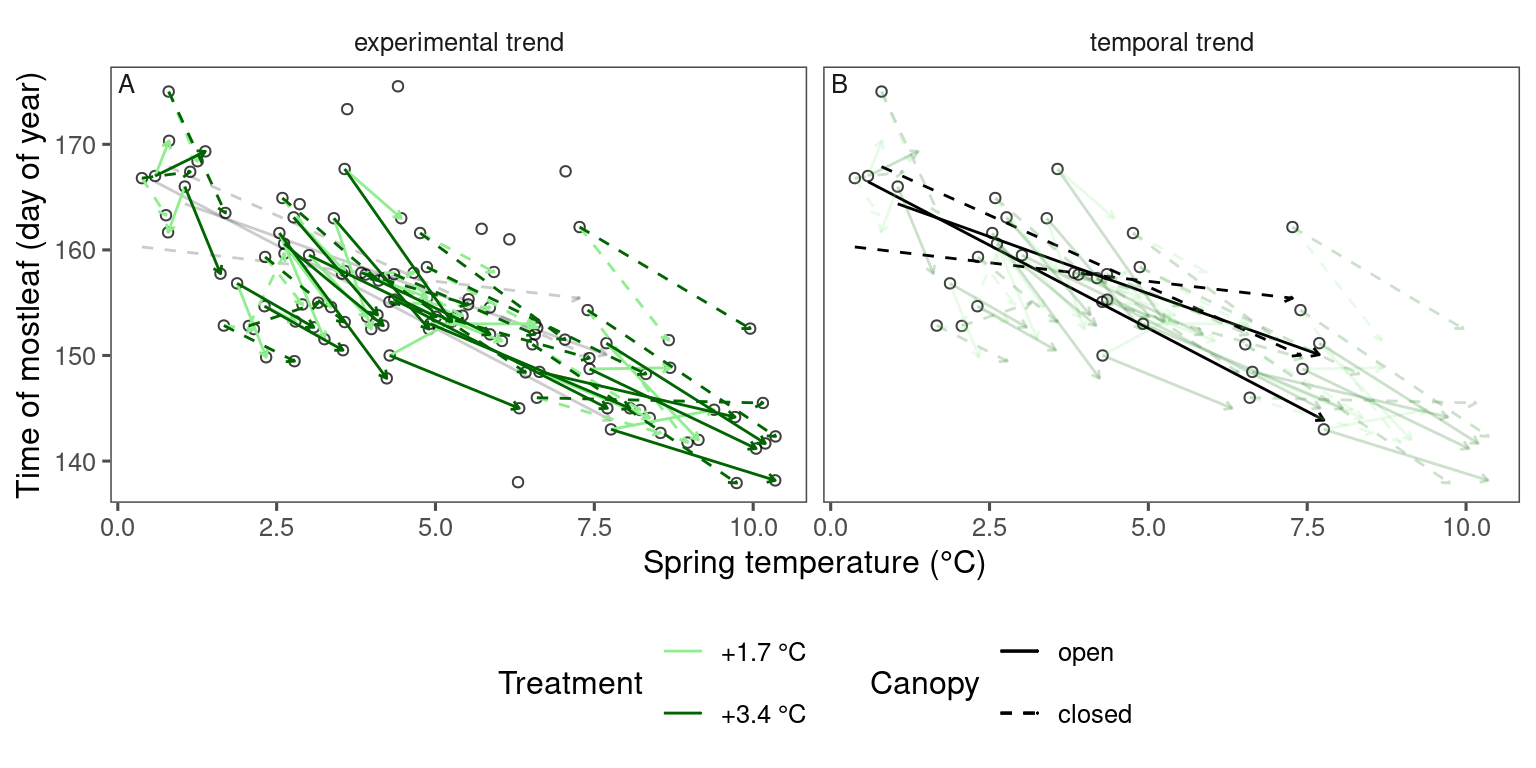
plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "senescence") %>%
filter(species == "acesa"),
dat_climate = dat_climate_fall,
var = "heat"
)
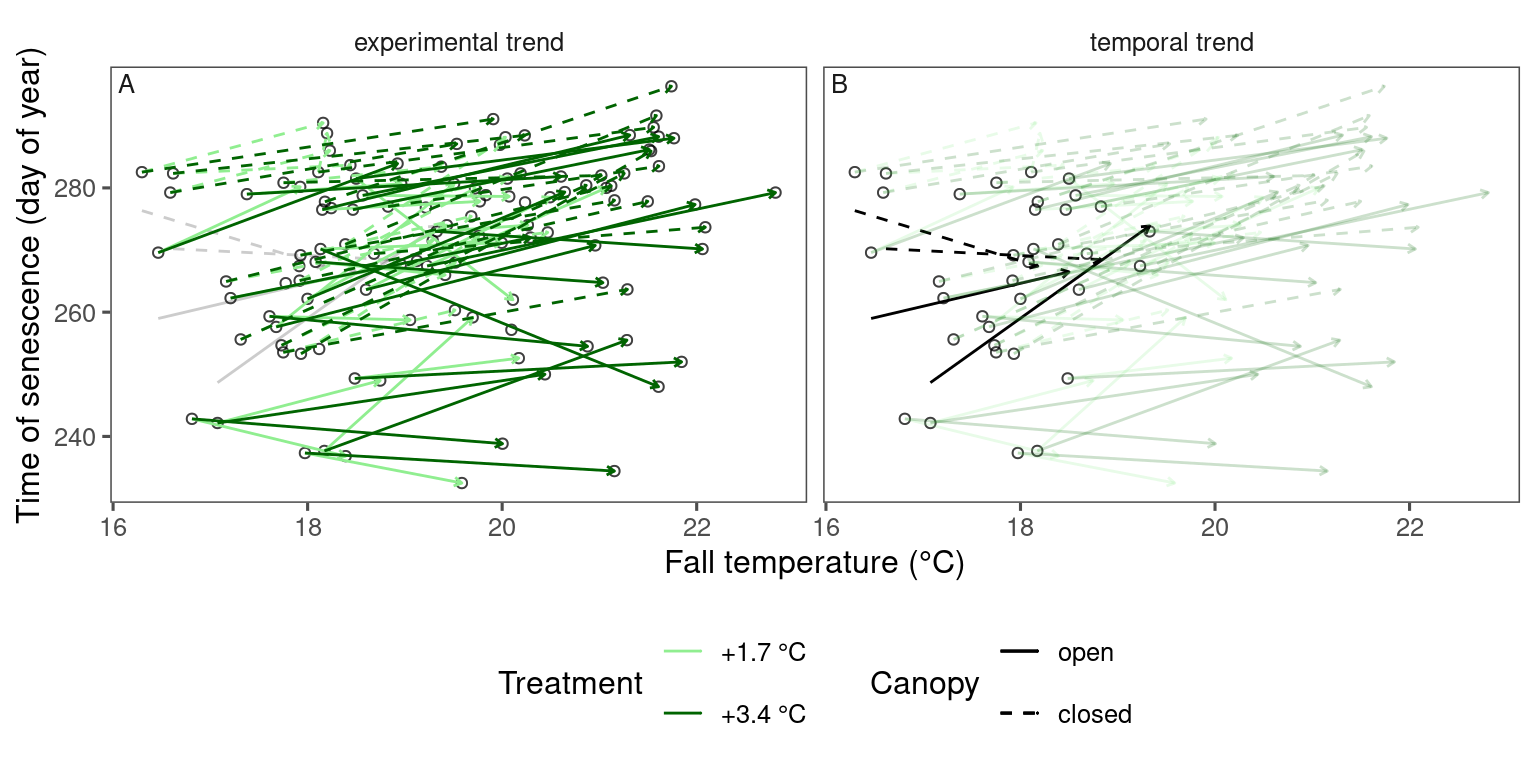
plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "leafdrop") %>%
filter(species == "pinst"),
dat_climate = dat_climate_fall,
var = "heat"
)
Plot comparisons
df_lme_spring <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "spring", full_factorial = T, derived = T, tidy_mcmc = T)
temp_sd_spring <- read_rds("alldata/intermediate/climate_spring_ambient.rds") %>%
attr("sd") %>%
`[`("temp")
df_compare_trends_spring <- test_compare_trends(df_lme_spring, temp_sd = temp_sd_spring) %>%
mutate(season = "spring") %>%
tidy_phenophase_name(season = "spring") %>%
tidy_model_name()
df_lme_fall <- read_bayes_all(path = "alldata/intermediate/phenophase/bg/", season = "fall", full_factorial = T, derived = T, tidy_mcmc = T)
temp_sd_fall <- read_rds("alldata/intermediate/climate_fall_ambient.rds") %>%
attr("sd") %>%
`[`("temp")
df_compare_trends_fall <- test_compare_trends(df_lme_fall, temp_sd = temp_sd_fall) %>%
mutate(season = "fall") %>%
tidy_phenophase_name(season = "fall")%>%
tidy_model_name()
df_compare_trends <- bind_rows(df_compare_trends_spring, df_compare_trends_fall) %>%
mutate(season = factor(season, levels = c("spring", "fall"))) %>%
tidy_species_name()
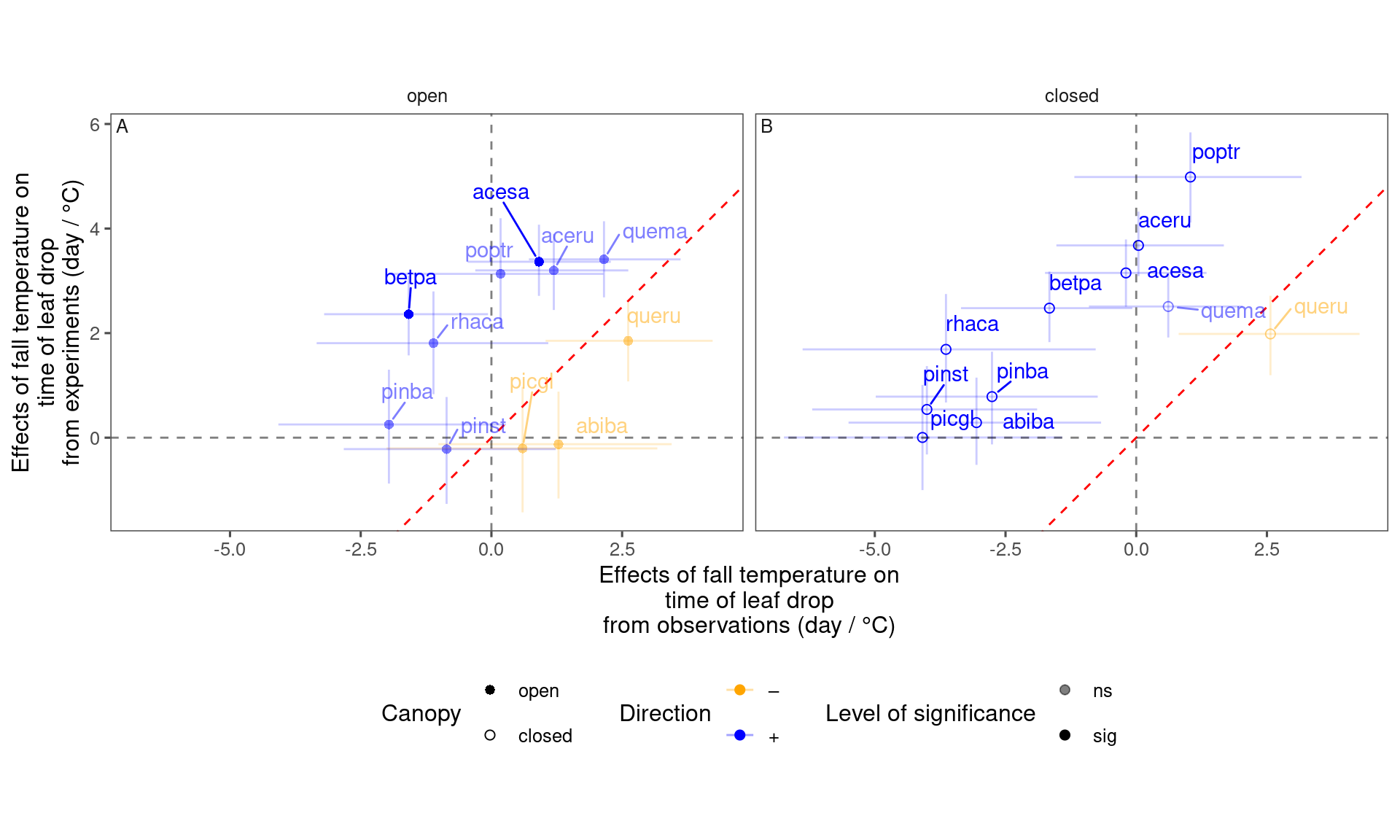
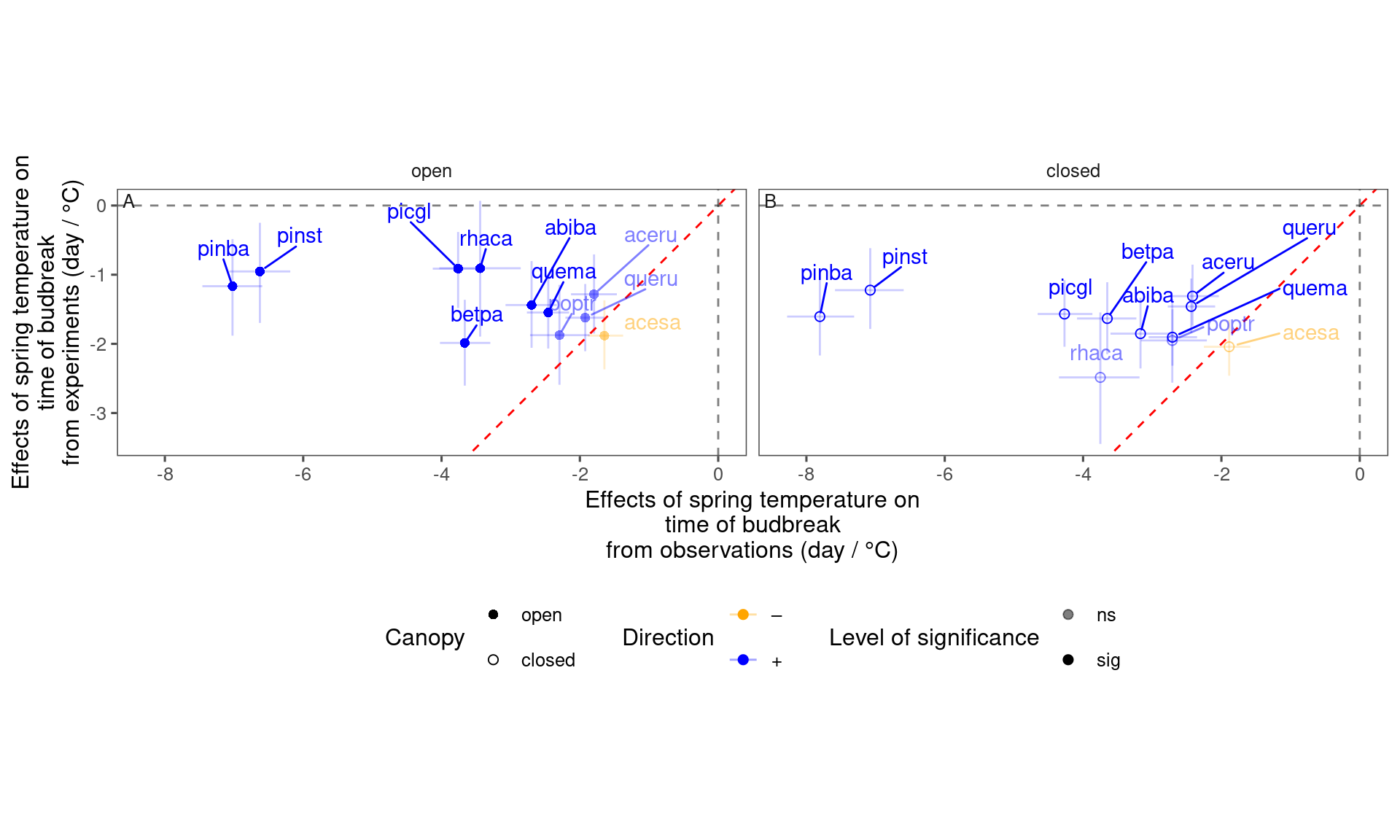
plot_trend_compare(df_compare_trends %>% filter(response == "start", season == "spring"), species_label = T)
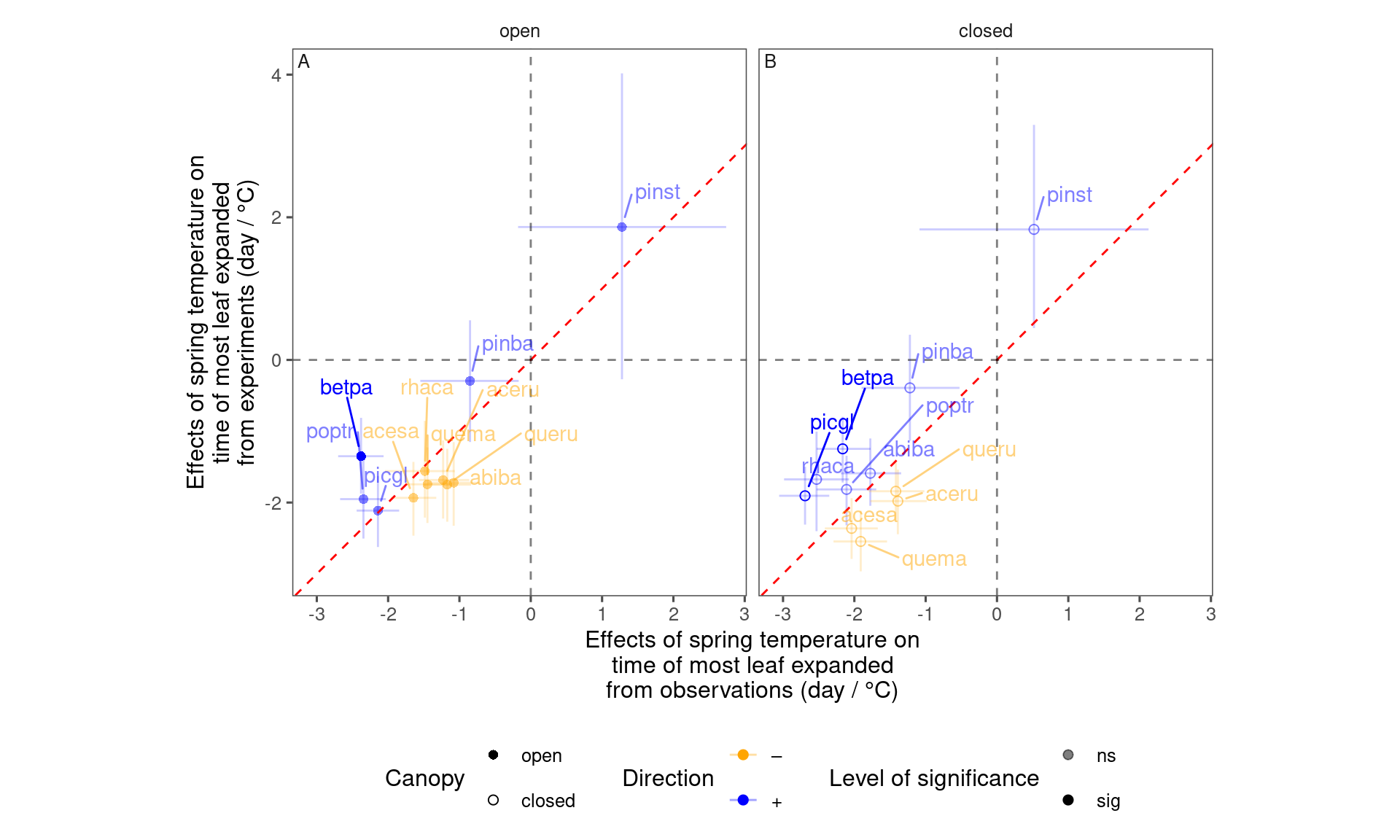
plot_trend_compare(df_compare_trends %>% filter(response == "end", season == "spring"), species_label = T)
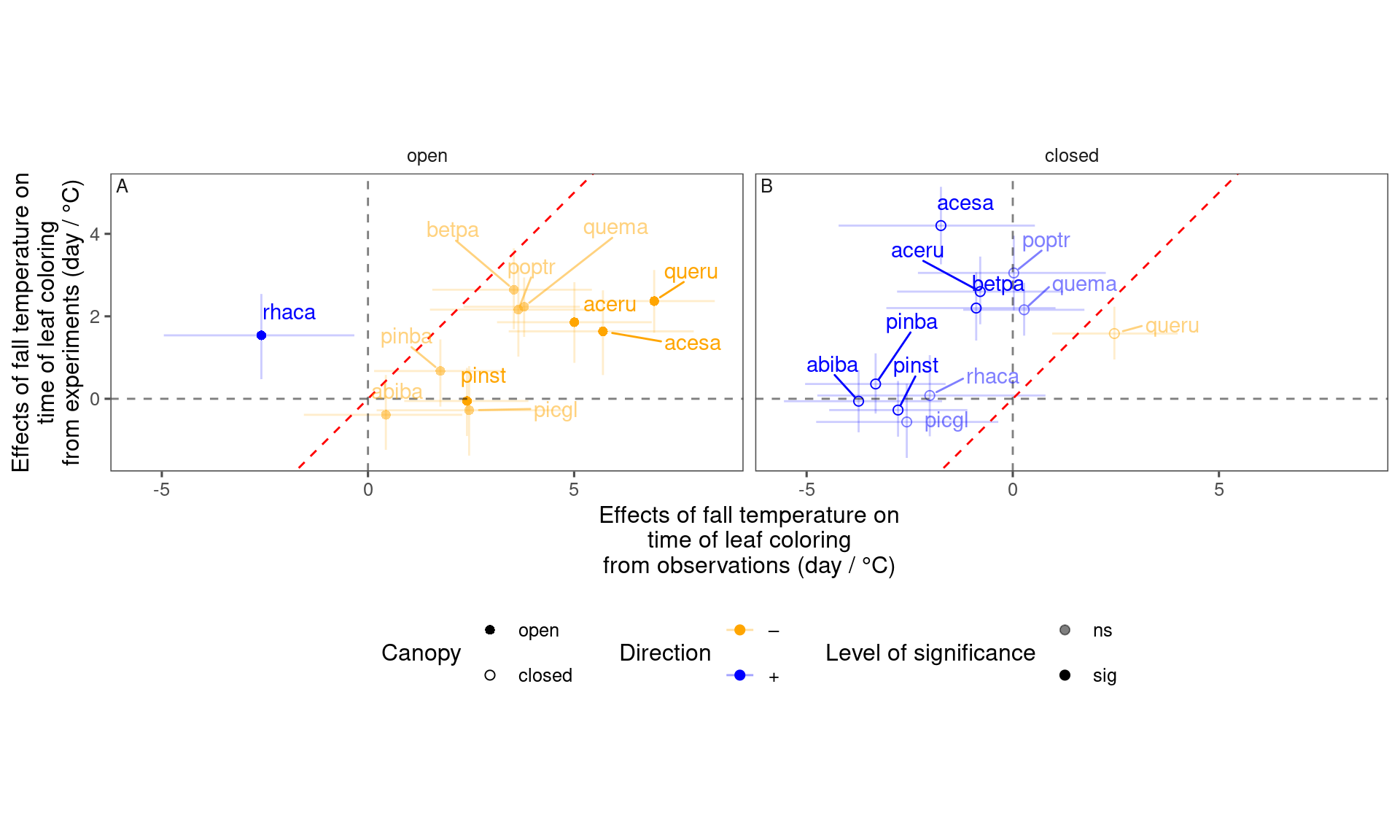
plot_trend_compare(df_compare_trends %>% filter(response == "start", season == "fall"), species_label = T)
plot_trend_compare(df_compare_trends %>% filter(response == "end", season == "fall"), species_label = T)