Our models for fall phenophases showed warming treatments generally delayed both the start and end of fall senescence (color change and leaf drop), often with no significant effects on the duration. Increase in background temperature delayed the start but not the end of fall senescence. These results are inconsistent with Zohner et al., 2023, which predicts that warming causes fall senescence to start earlier and end later. Our results did not point to clear sink-driven advancement of senescence under warming.
In order to test the sink-driven hypothesis, I explore the relationship between spring and fall phenophases.
Calculate phenophase time.
dat_phenophase_time_clean %>%
group_by(barcode, year, phenophase, doy) %>%
filter(n() > 1) %>%
ungroup() %>%
arrange(barcode, year, phenophase, doy)
## # A tibble: 38 × 15
## site canopy heat heat_name water water_name block plot species barcode
## <chr> <chr> <fct> <fct> <fct> <fct> <chr> <chr> <chr> <dbl>
## 1 cfc closed _ ambient _ ambient a a5 acene 31469
## 2 cfc closed _ ambient _ ambient a a5 tilam 31469
## 3 cfc closed _ ambient _ ambient a a5 aceru 31469
## 4 cfc closed _ ambient _ ambient a a5 tilam 31469
## 5 cfc closed _ ambient _ ambient a a3 tsuca 31554
## 6 cfc closed + +1.7C _ ambient a a6 pinre 31554
## 7 cfc closed _ ambient _ ambient a a3 tsuca 31554
## 8 cfc closed + +1.7C _ ambient a a6 pinre 31554
## 9 cfc closed _ ambient _ ambient a a3 tsuca 31554
## 10 cfc closed + +1.7C _ ambient a a6 pinre 31554
## # ℹ 28 more rows
## # ℹ 5 more variables: cohort <dbl>, year <dbl>, doy <dbl>, phenophase <fct>,
## # notes <chr>
Found these probelamtic data entries.
dat_phenophase_wide <- dat_phenophase_time_clean %>%
filter(phenophase != "oneleaf") %>%
select(species, barcode, year, phenophase, doy) %>%
distinct(species, barcode, year, phenophase, .keep_all = T) %>% # this step would not be necessary if the errors in data are resolved.
pivot_wider(names_from = phenophase, values_from = doy) %>%
drop_na() %>%
pivot_longer(cols = budbreak:mostleaf, names_to = "spring", values_to = "doy")
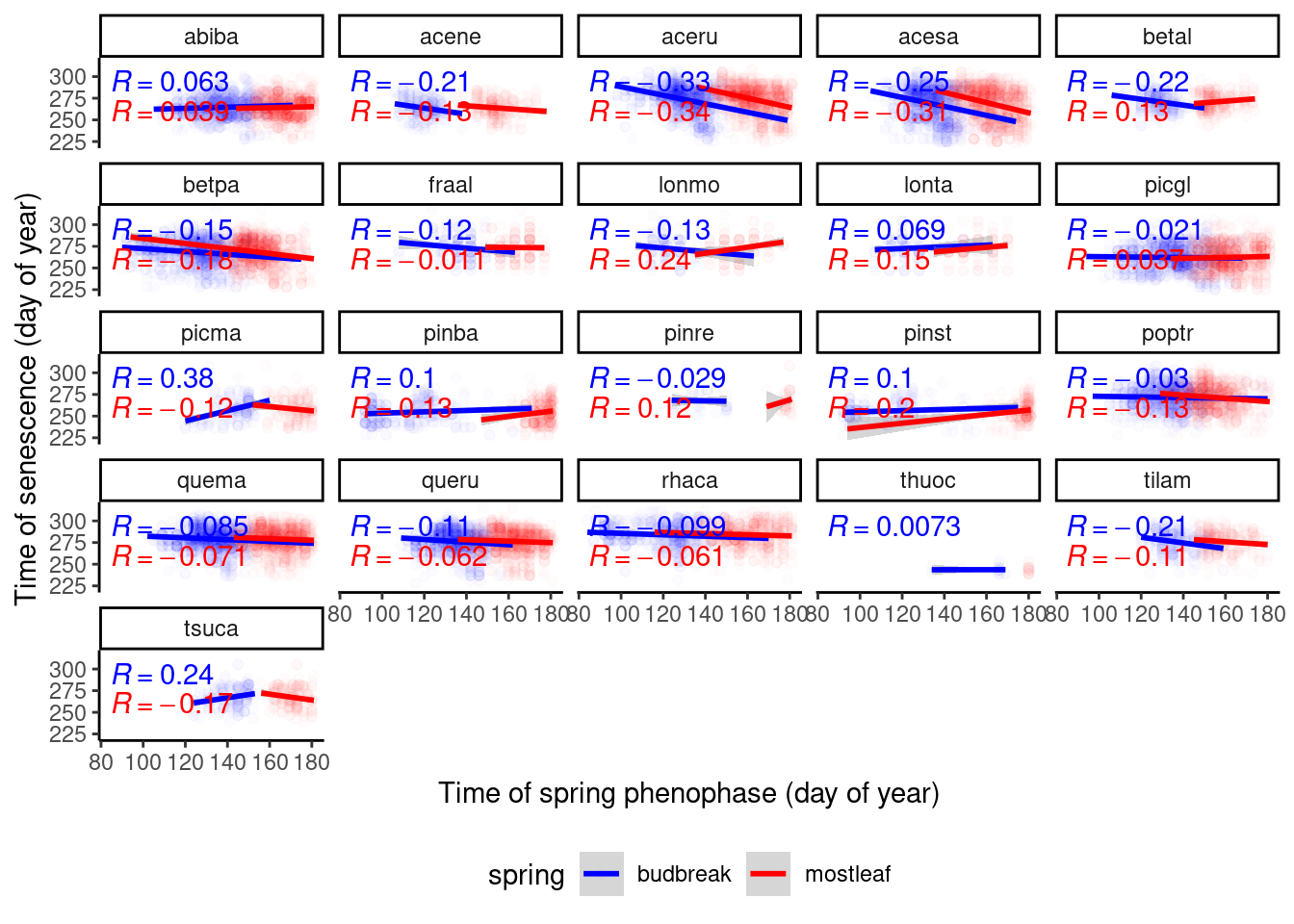
Exploratory plot.
dat_phenophase_wide %>%
ggplot() +
geom_point(aes(x = doy, y = senescence, col = spring), alpha = 0.01) +
geom_smooth(aes(x = doy, y = senescence, col = spring), method = "lm") +
ggpubr::stat_cor(aes(x = doy, y = senescence, group = spring, col = spring, label = after_stat(r.label)), show.legend = F) +
scale_color_manual(values = c("budbreak" = "blue", "mostleaf" = "red")) +
facet_wrap(. ~ species) +
labs(
x = "Time of spring phenophase (day of year)",
y = "Time of senescence (day of year)"
) +
theme_classic() +
theme(legend.position = "bottom")
dat_phenophase_wide %>%
ggplot() +
geom_point(aes(x = doy, y = leafdrop, col = spring), alpha = 0.01) +
geom_smooth(aes(x = doy, y = leafdrop, col = spring), method = "lm") +
ggpubr::stat_cor(aes(x = doy, y = leafdrop, group = spring, col = spring, label = after_stat(r.label)), show.legend = F) +
scale_color_manual(values = c("budbreak" = "blue", "mostleaf" = "red")) +
facet_wrap(. ~ species) +
labs(
x = "Time of spring phenophase (day of year)",
y = "Time of leafdrop (day of year)"
) +
theme_classic() +
theme(legend.position = "bottom")
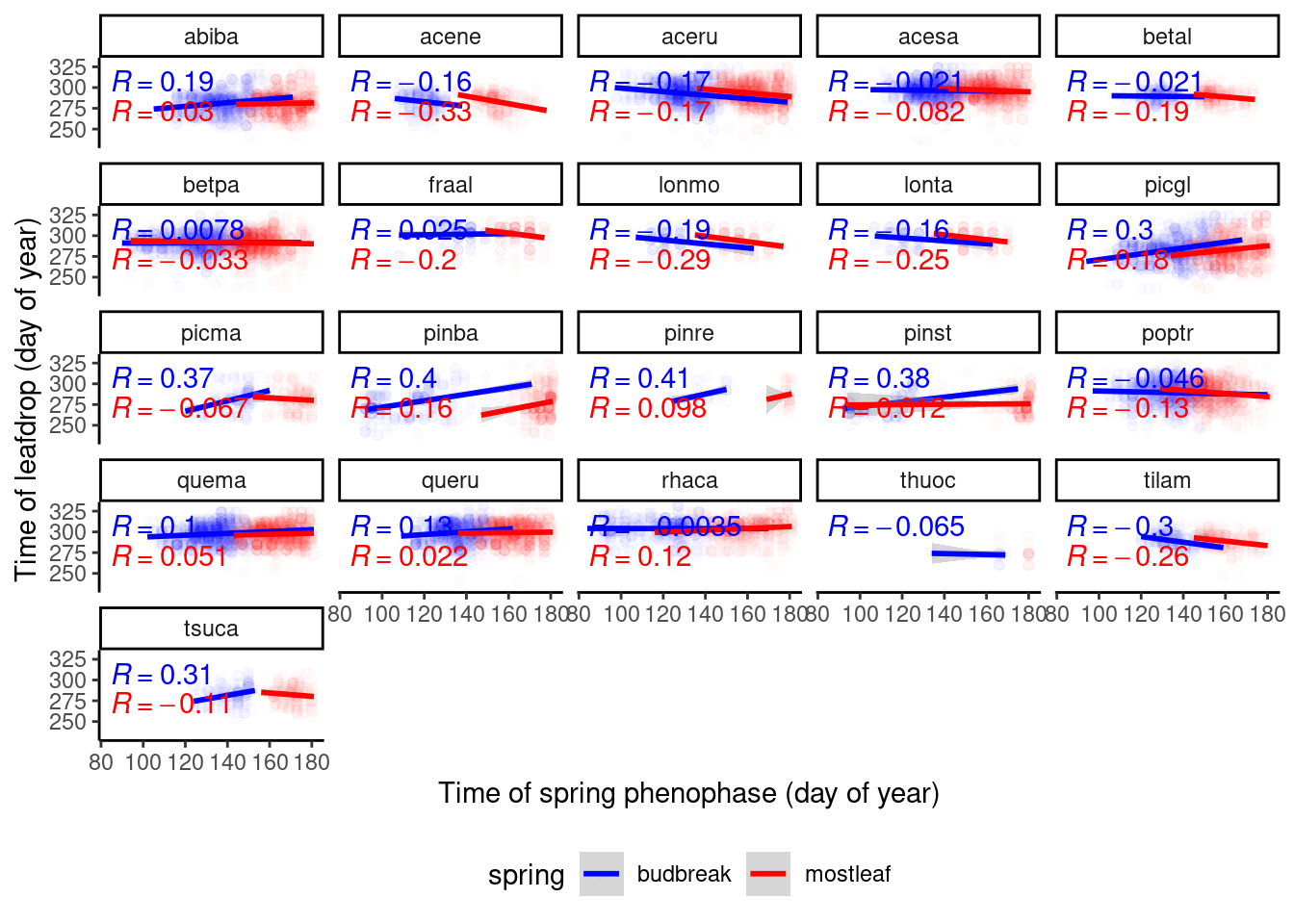
If fall phenophases are limited by sink, we would expect positive correlations between spring and fall phenophases (earlier spring events - earlier fall events). There did not seem to be strong positive correlation. This does not rule out the possibility that limitation by sink attenutates possible delay driven by warming. I did not proceed to fit Bayesian LME.