Effects by species and treatments
Make a synthesis figure combining shoot and phenophase responses to treatments.
species <- "queru"
df_shoot <- read_rds(str_c("alldata/intermediate/shootmodeling/uni/", species, "/df_data.rds"))
design <- tidy_species_design(df_shoot)
df_shoot_coef <- read_rds(str_c("alldata/intermediate/shootmodeling/uni/", species, "/df_MCMC.rds")) %>%
tidy_mcmc(df_shoot) %>%
summ_mcmc(option = "all", stats = "median") %>%
filter(is.na(group)) %>%
select(-group)
df_shoot_pred <- read_rds(str_c("alldata/intermediate/shootmodeling/uni/", species, "/df_pred_marginal.rds"))
df_phenophase_spring <- read_rds(str_c("alldata/intermediate/phenophase/uni/", species, "/spring/df_data.rds"))
df_phenophase_spring_coef <- read_rds(str_c("alldata/intermediate/phenophase/uni/", species, "/spring/df_MCMC.rds")) %>%
tidy_mcmc(df_phenophase_spring) %>%
summ_mcmc(option = "all", stats = "median")
df_phenophase_fall <- read_rds(str_c("alldata/intermediate/phenophase/uni/", species, "/fall/df_data.rds"))
df_phenophase_fall_coef <- read_rds(str_c("alldata/intermediate/phenophase/uni/", species, "/fall/df_MCMC.rds")) %>%
tidy_mcmc(df_phenophase_fall) %>%
summ_mcmc(option = "all", stats = "median")
(p_warming <- plot_synthesis_one_trt(df_shoot_pred, df_shoot_coef, df_phenophase_spring_coef, df_phenophase_fall_coef, treatment = "warming", spring_only = T))
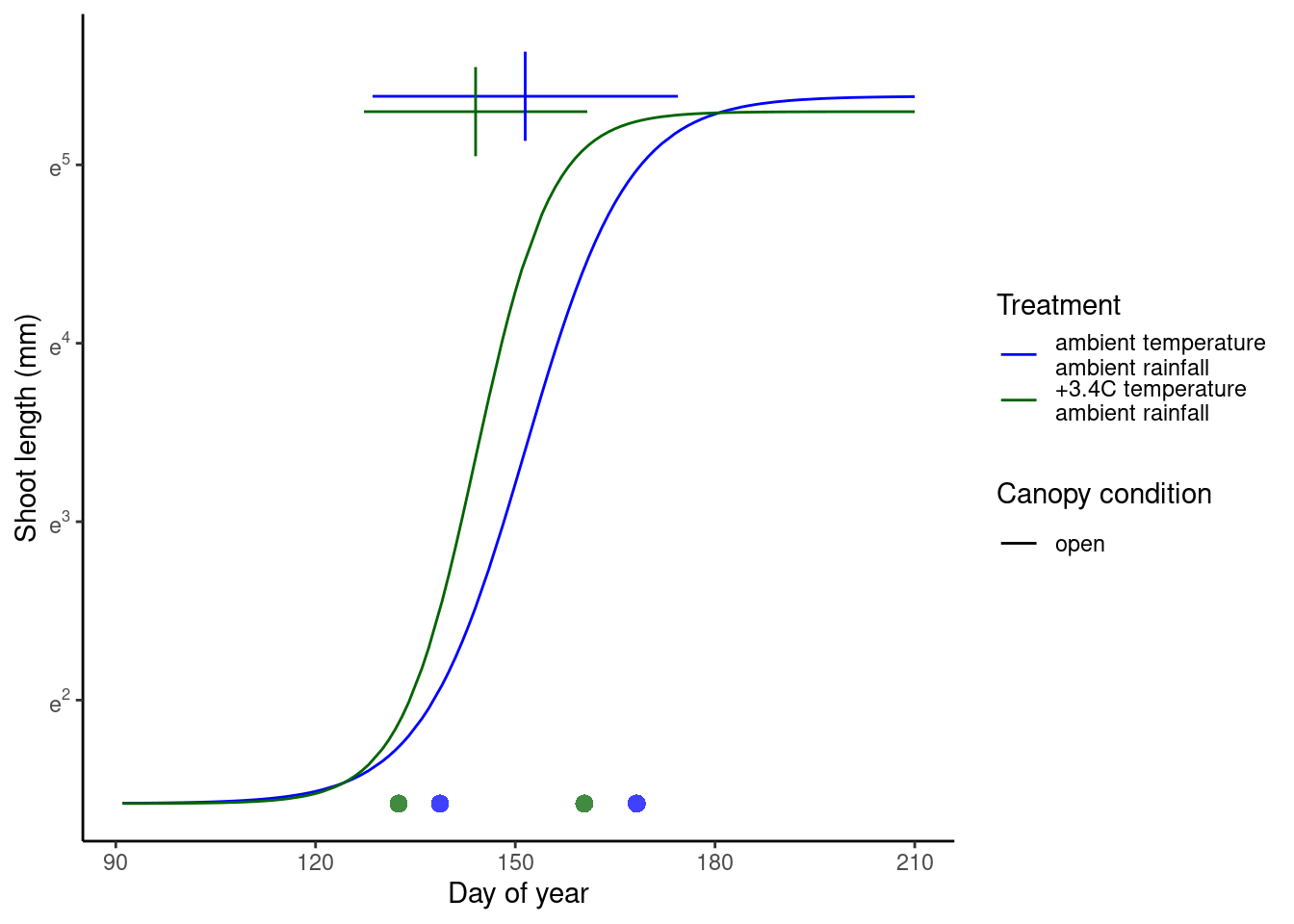
(p_drying <- plot_synthesis_one_trt(df_shoot_pred, df_shoot_coef, df_phenophase_spring_coef, df_phenophase_fall_coef, treatment = "drying", spring_only = T))
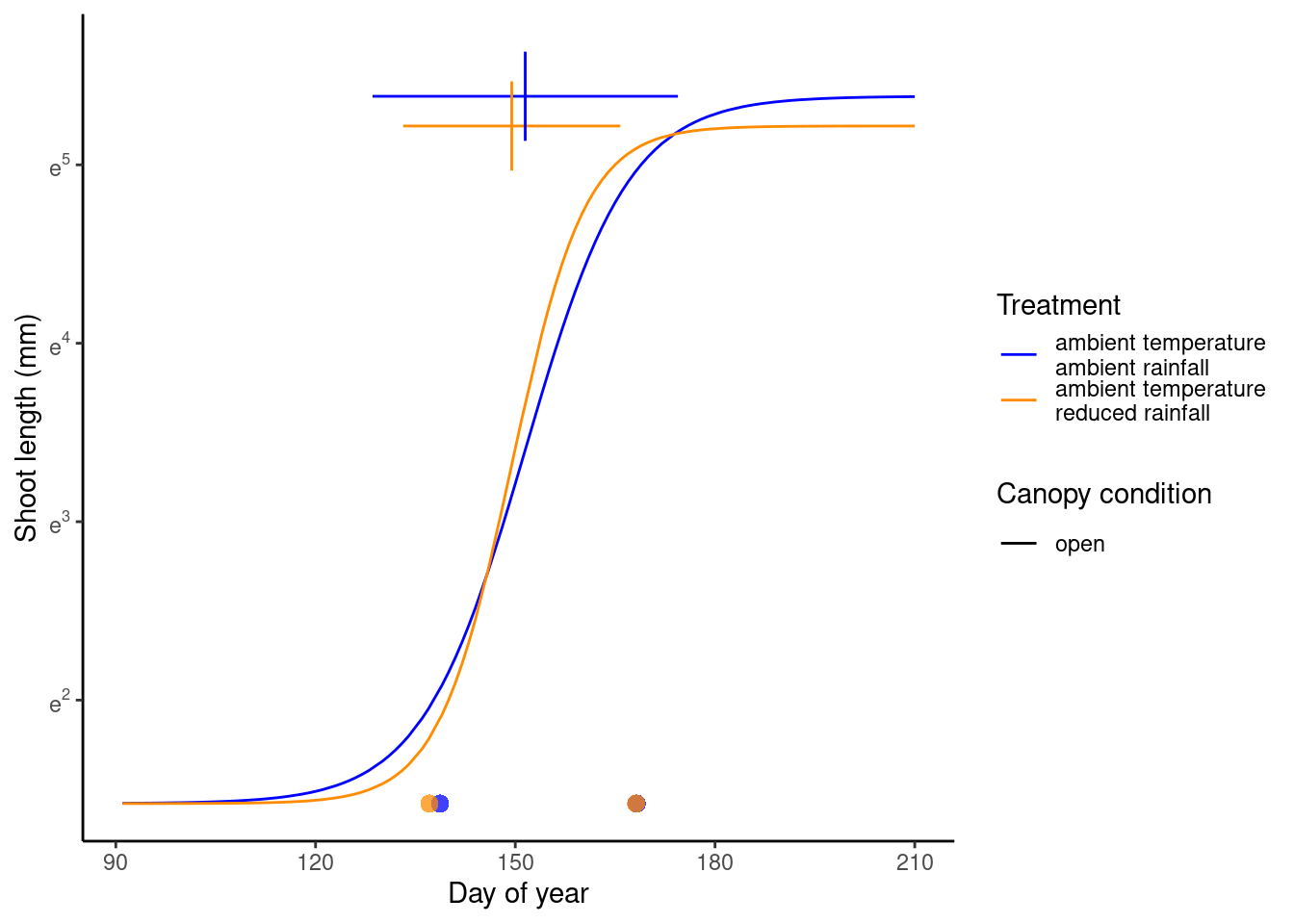
(p_closed <- plot_synthesis_one_trt(df_shoot_pred, df_shoot_coef, df_phenophase_spring_coef, df_phenophase_fall_coef, treatment = "closed", spring_only = T))
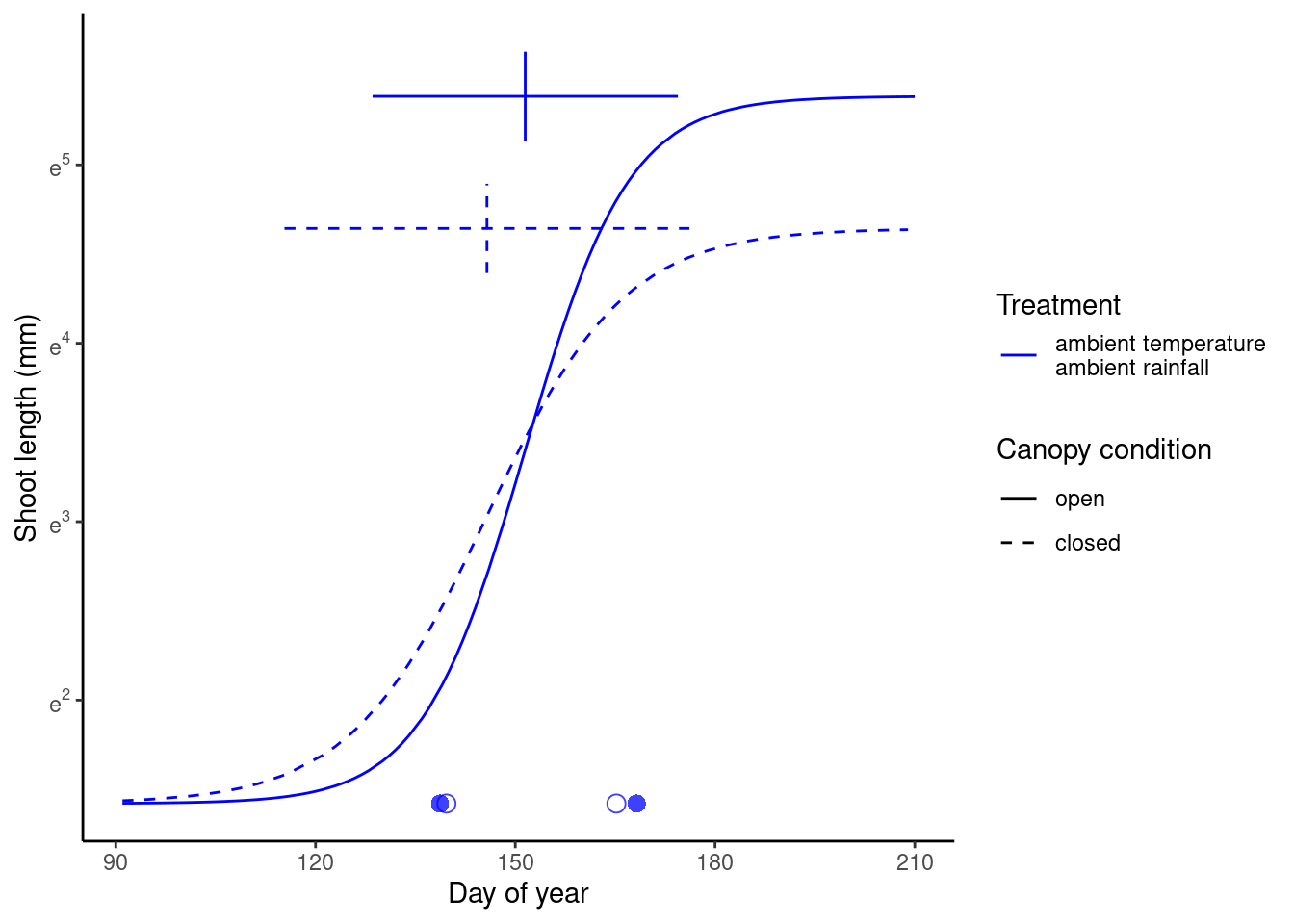
plot_synthesis(v_species <- dat_shoot %>% pull(species) %>% unique(), spring_only = T, path = "alldata/output/figures/synthesis/")
Effects across species for each treatment
Make an ordination plot to see patterns of responses in phenophase and shoot growth to a treatment.
df_coef_shoot <- read_bayes_all(path = "alldata/intermediate/shootmodeling/uni/", full_factorial = T, derived = F) %>%
filter(!is.na(covariate)) %>%
summ_mcmc(option = "coef", stats = "median")
df_coef_phenophase <- read_bayes_all(path = "alldata/intermediate/phenophase/uni/", season = "spring", full_factorial = T, derived = F) %>%
filter(!is.na(covariate)) %>%
summ_mcmc(option = "coef", stats = "median")
plot_ordination(df_coef_shoot, df_coef_phenophase, treatment = "warming")
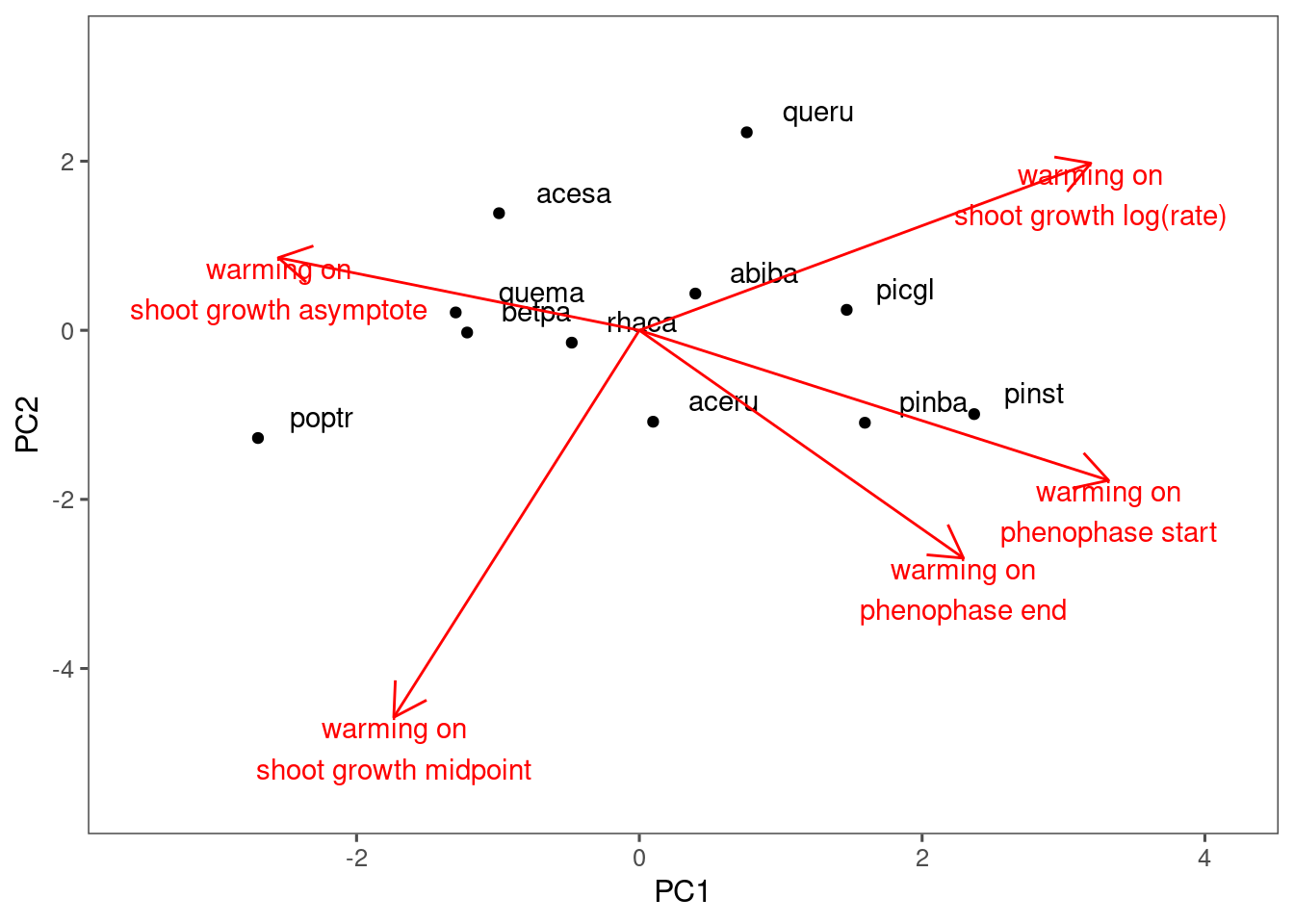 Differentiated mostly by the responses in asymptote. Not much difference in the responses in timings.
Differentiated mostly by the responses in asymptote. Not much difference in the responses in timings.
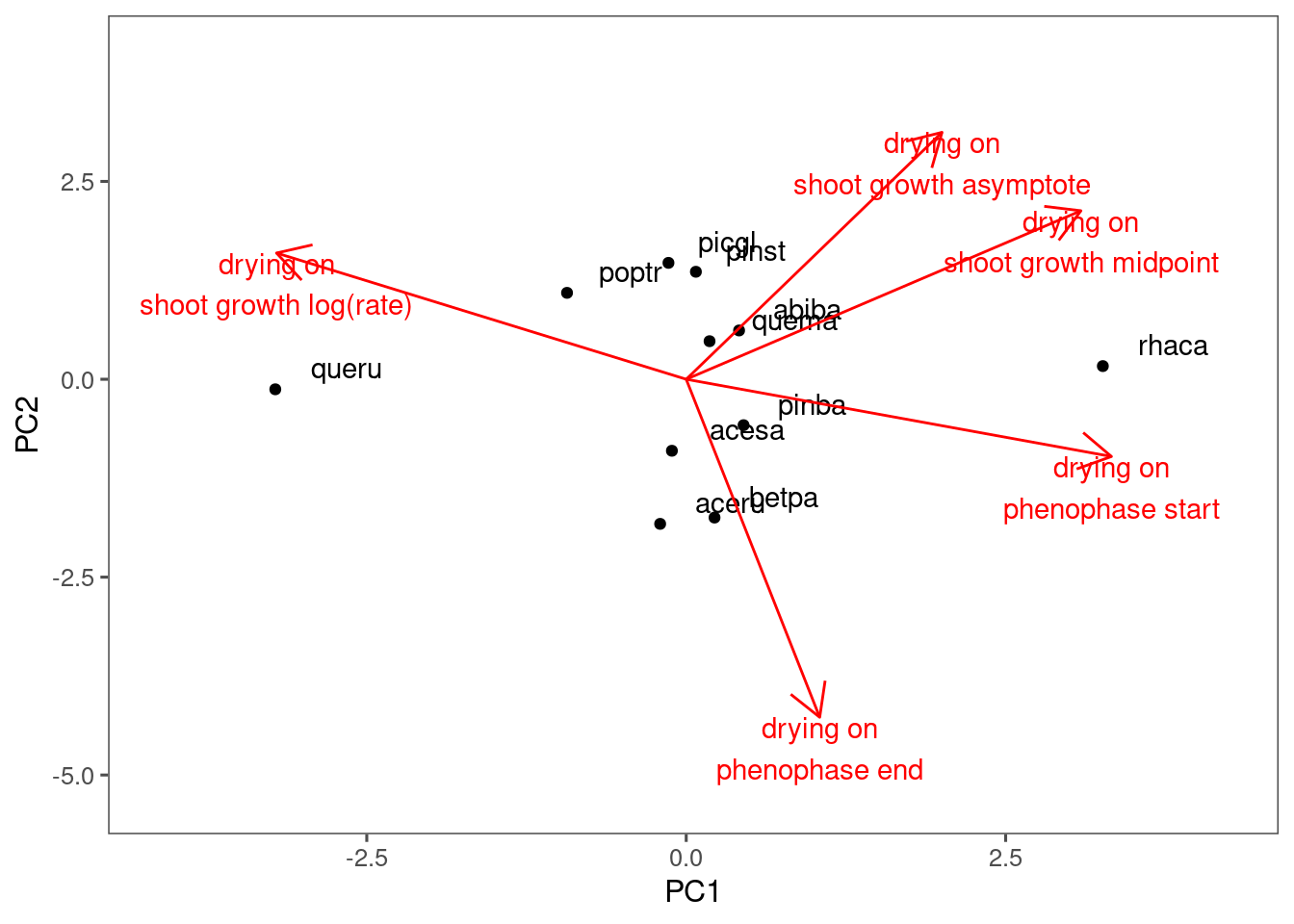
plot_ordination(df_coef_shoot, df_coef_phenophase, treatment = "drying")
plot_ordination(df_coef_shoot, df_coef_phenophase, treatment = "closed")
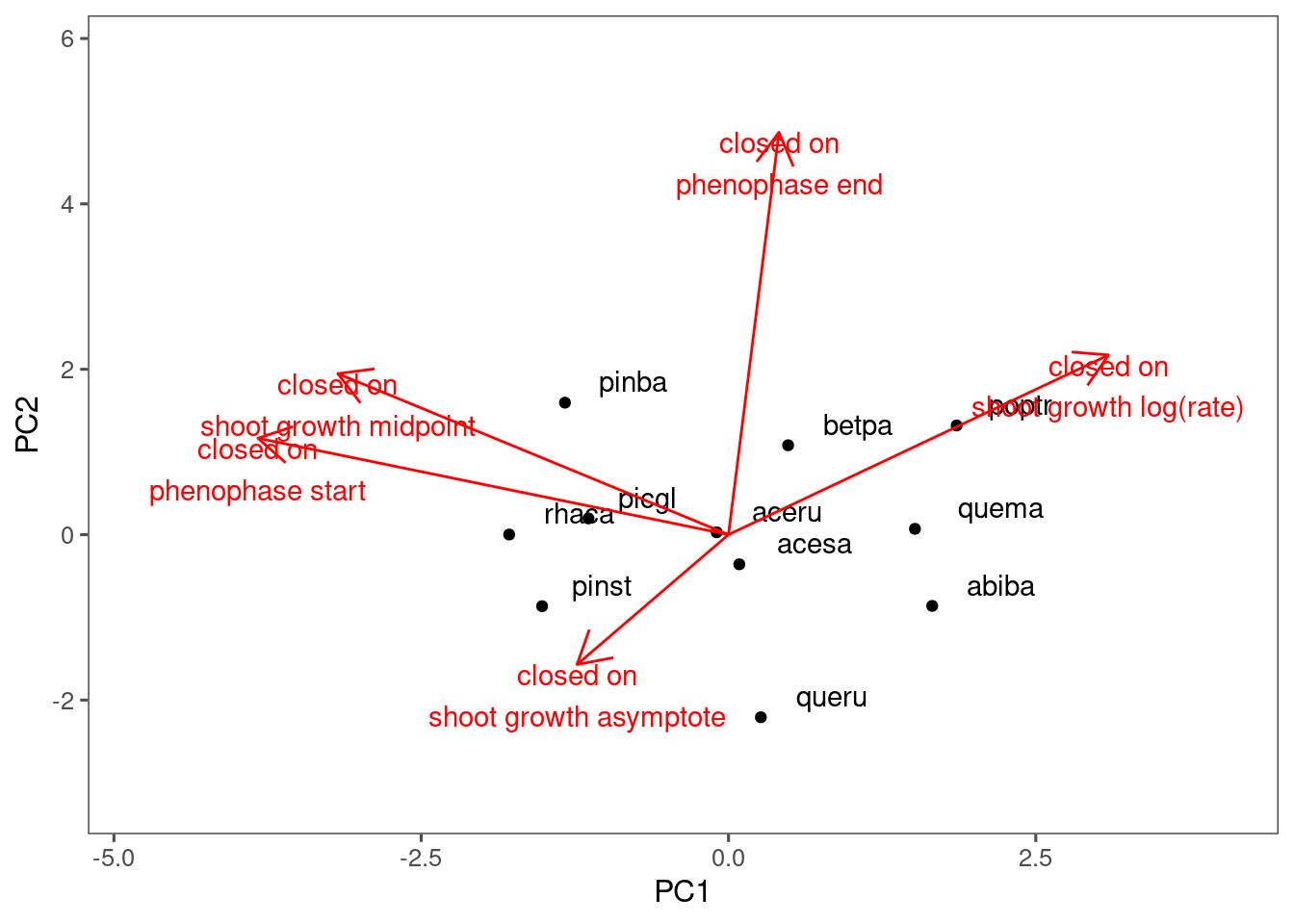
I did not see much clustering in the responses to drying or closed canpopy.