Prepare data
dat_12 <- dat_shoot_cov %>%
filter(species == "queru") %>%
filter(shoot > 0) %>%
filter(doy > 90, doy <= 210) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = str_c(site, canopy, barcode, slot, year, sep = "_") %>% factor() %>% as.integer())
Fit model
df_MCMC_12 <- calc_bayes_fit(
data = dat_12 %>% mutate(tag = "training"),
version = 12
)
write_rds(df_MCMC_12, "alldata/intermediate/shootmodeling/df_MCMC_12.rds")
df_MCMC_12 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_12.rds")
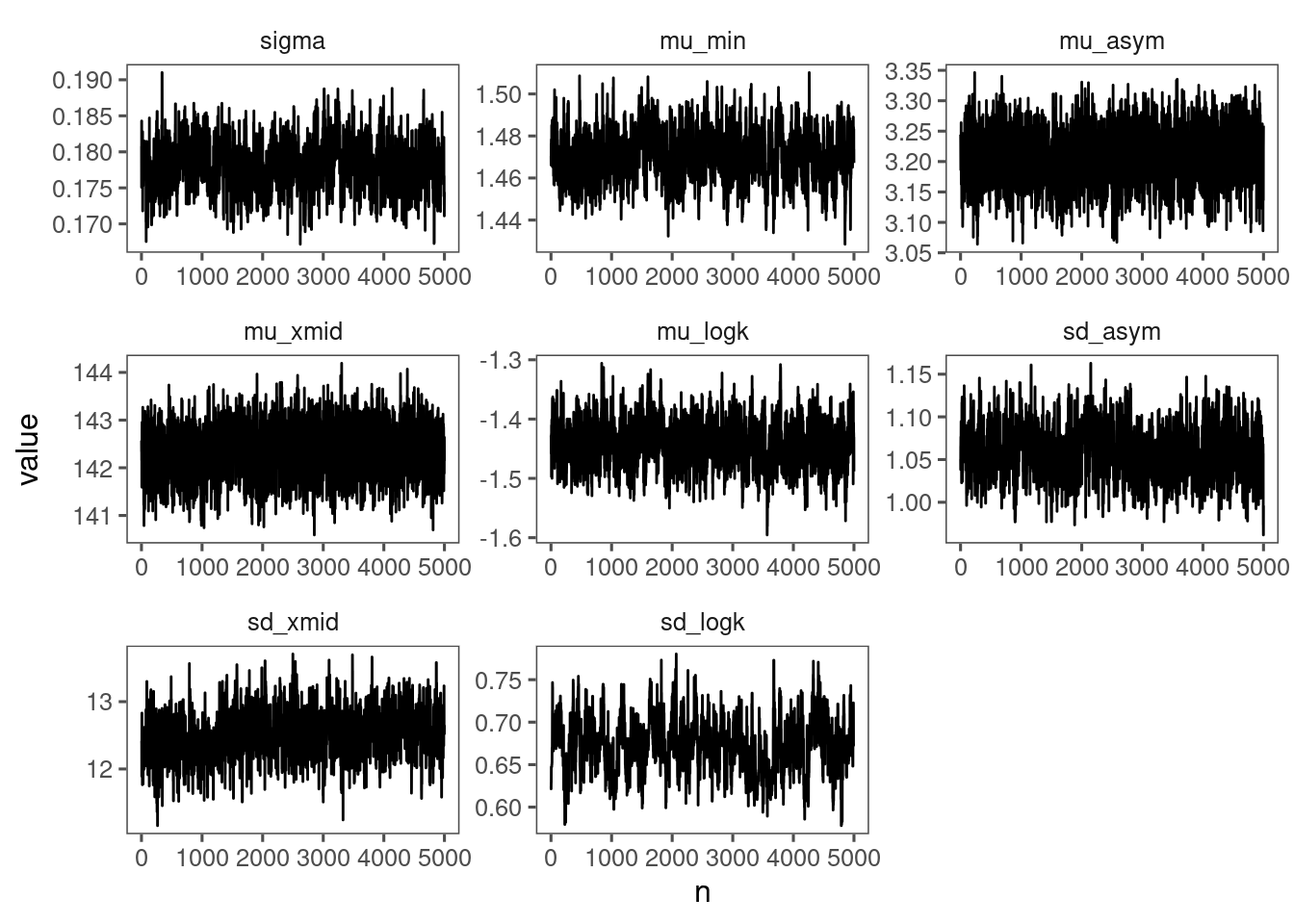
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_12)
p_bayes_diagnostics$p_MCMC
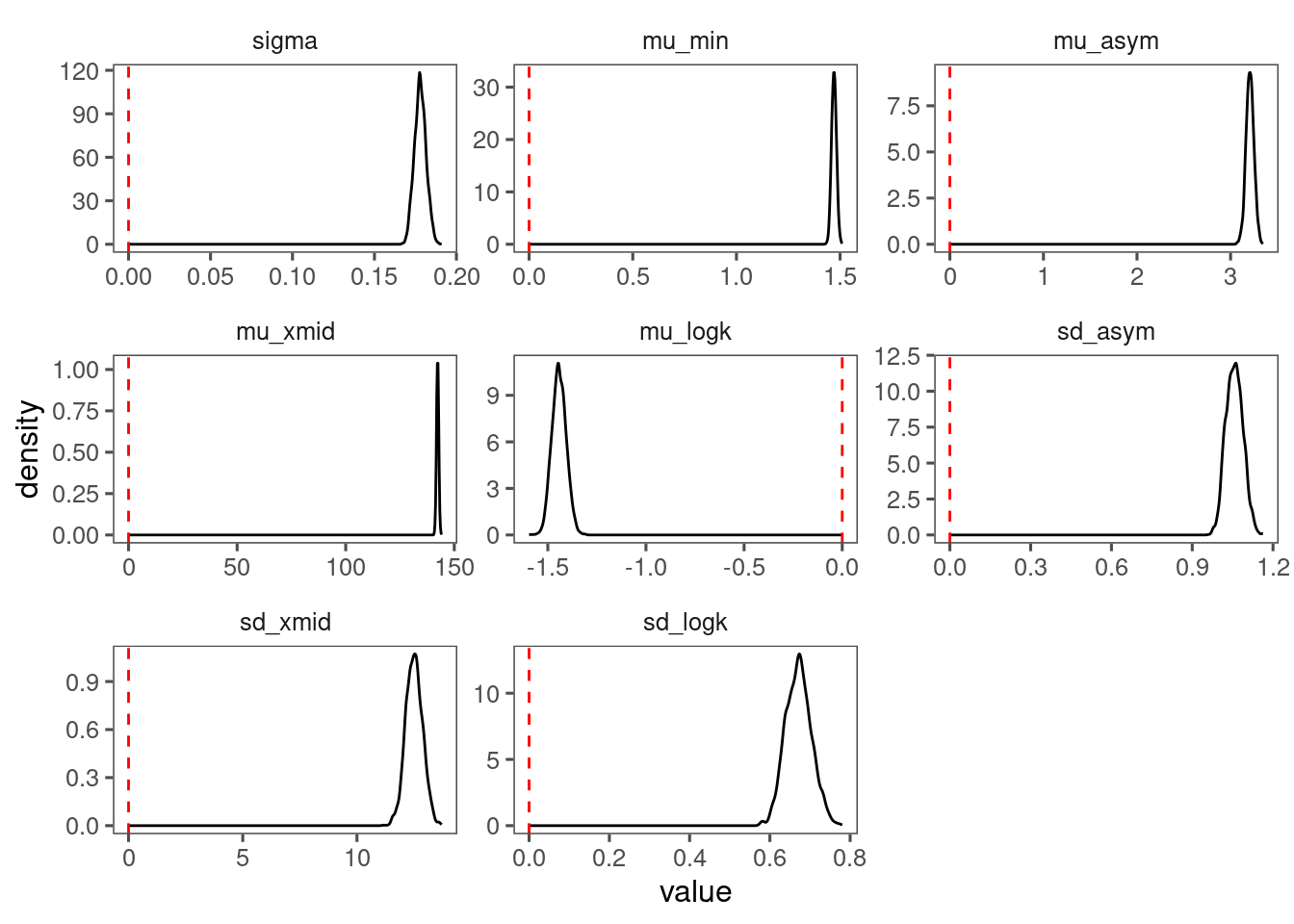
p_bayes_diagnostics$p_posterior
Make predictions
df_pred_12 <- calc_bayes_predict(
data = dat_12 %>%
distinct(group, doy),
df_MCMC = df_MCMC_12,
version = 12
)
write_rds(df_pred_12, "alldata/intermediate/shootmodeling/df_pred_12.rds")
df_pred_12 <- read_rds("alldata/intermediate/shootmodeling/df_pred_12.rds")
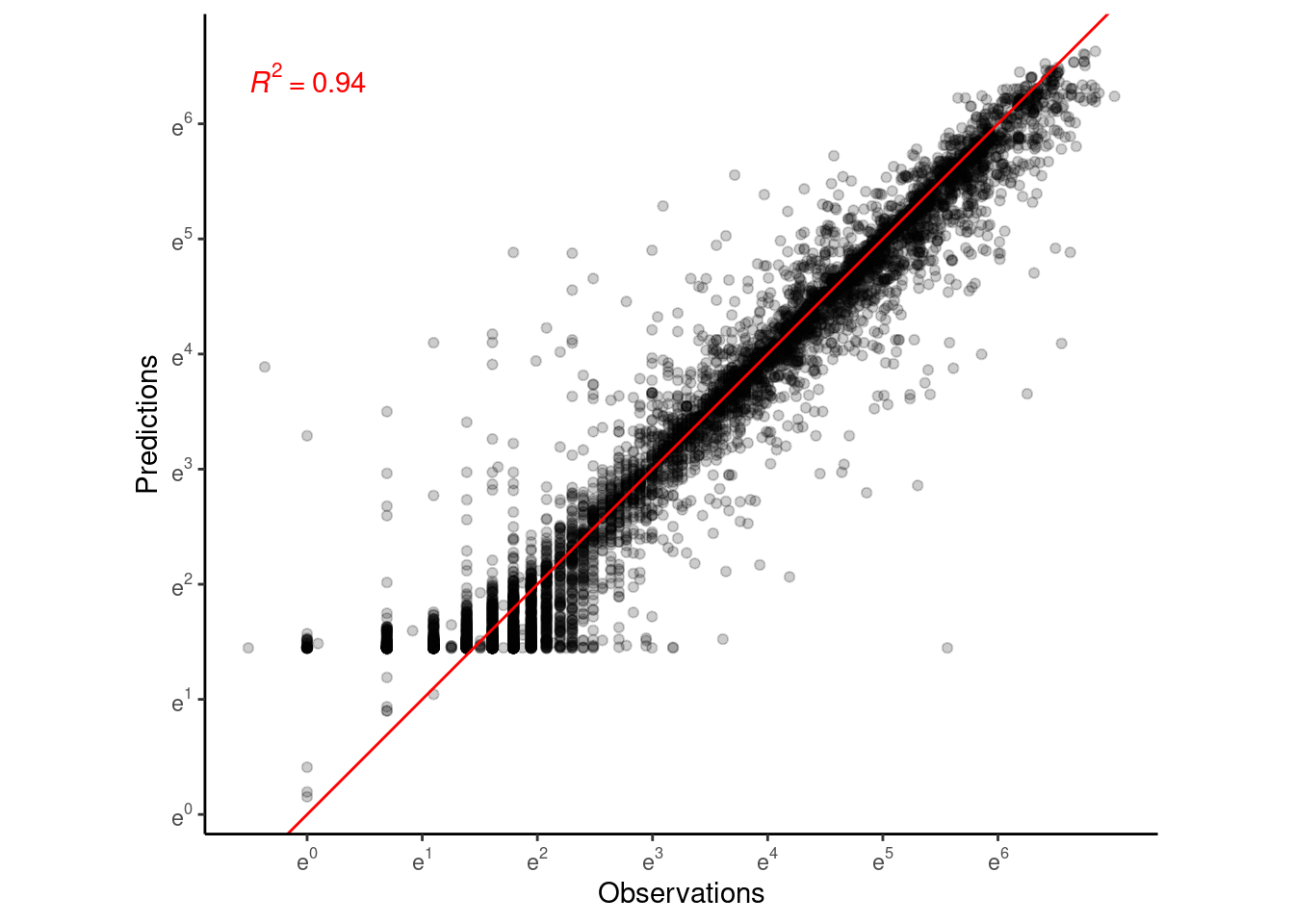
p_bayes_predict <- plot_bayes_predict(
data = dat_12,
data_predict = df_pred_12,
vis_log = T,
vis_ci = F
)
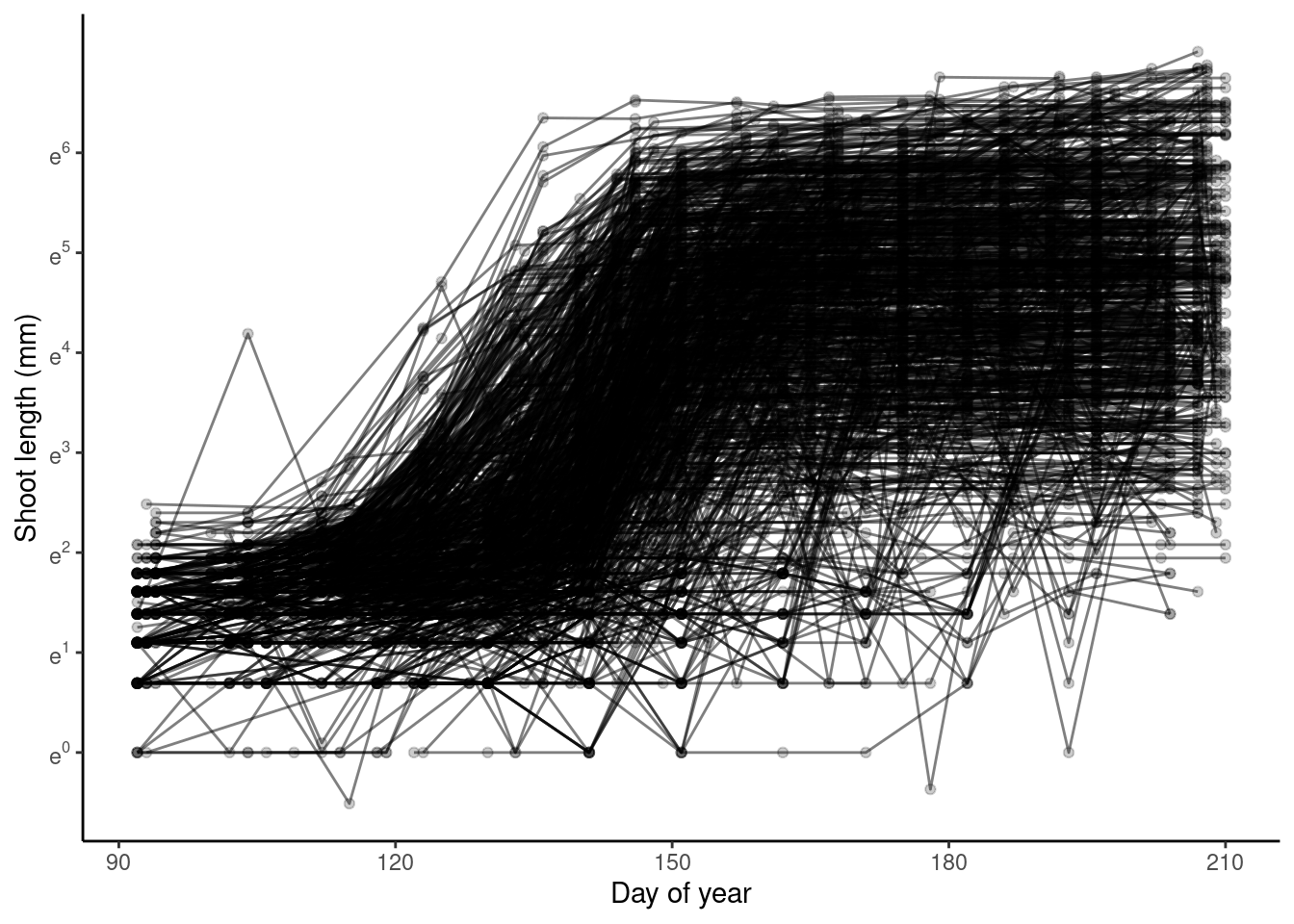
p_bayes_predict$p_original
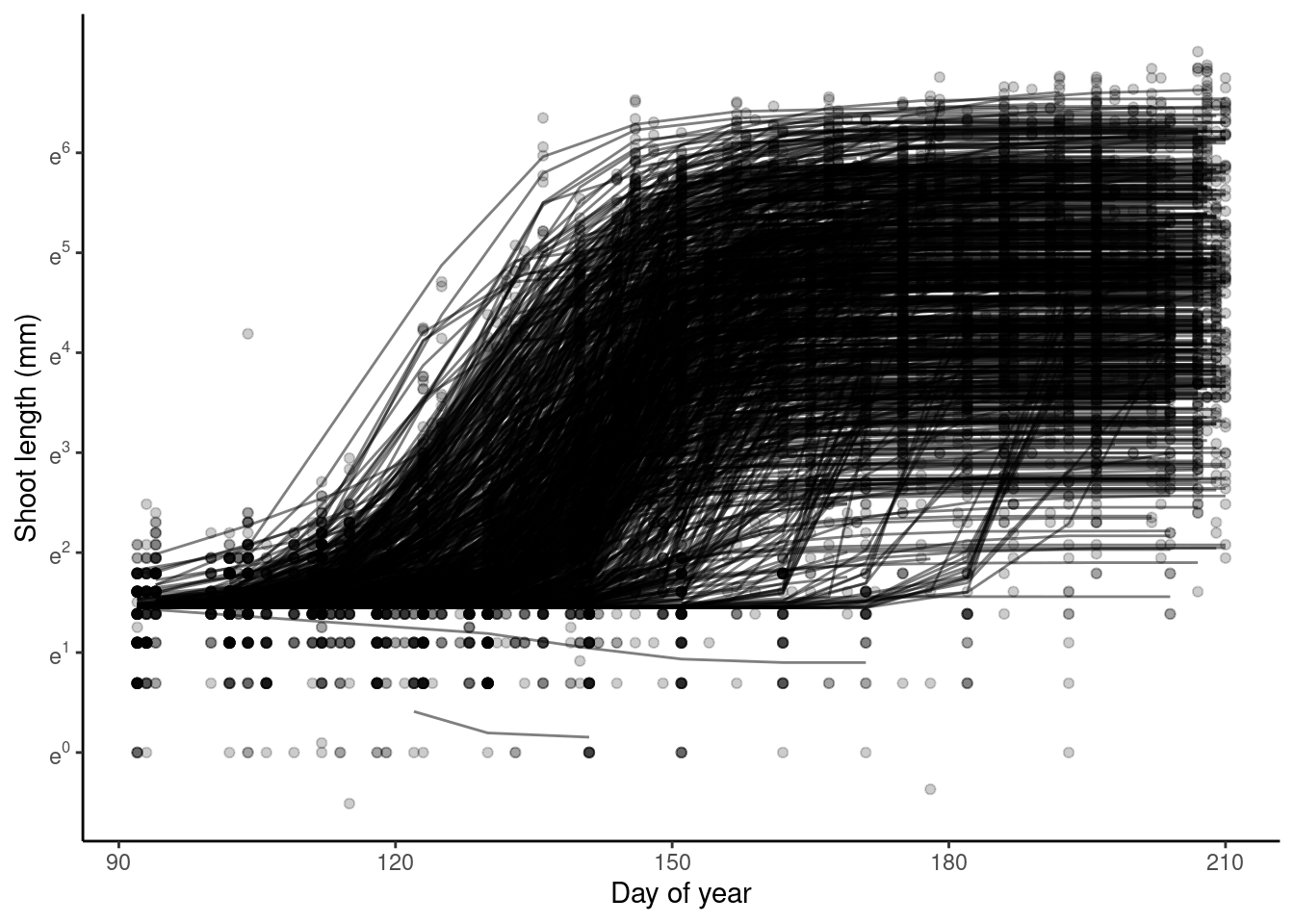
p_bayes_predict$p_overlay
p_bayes_predict$p_accuracy
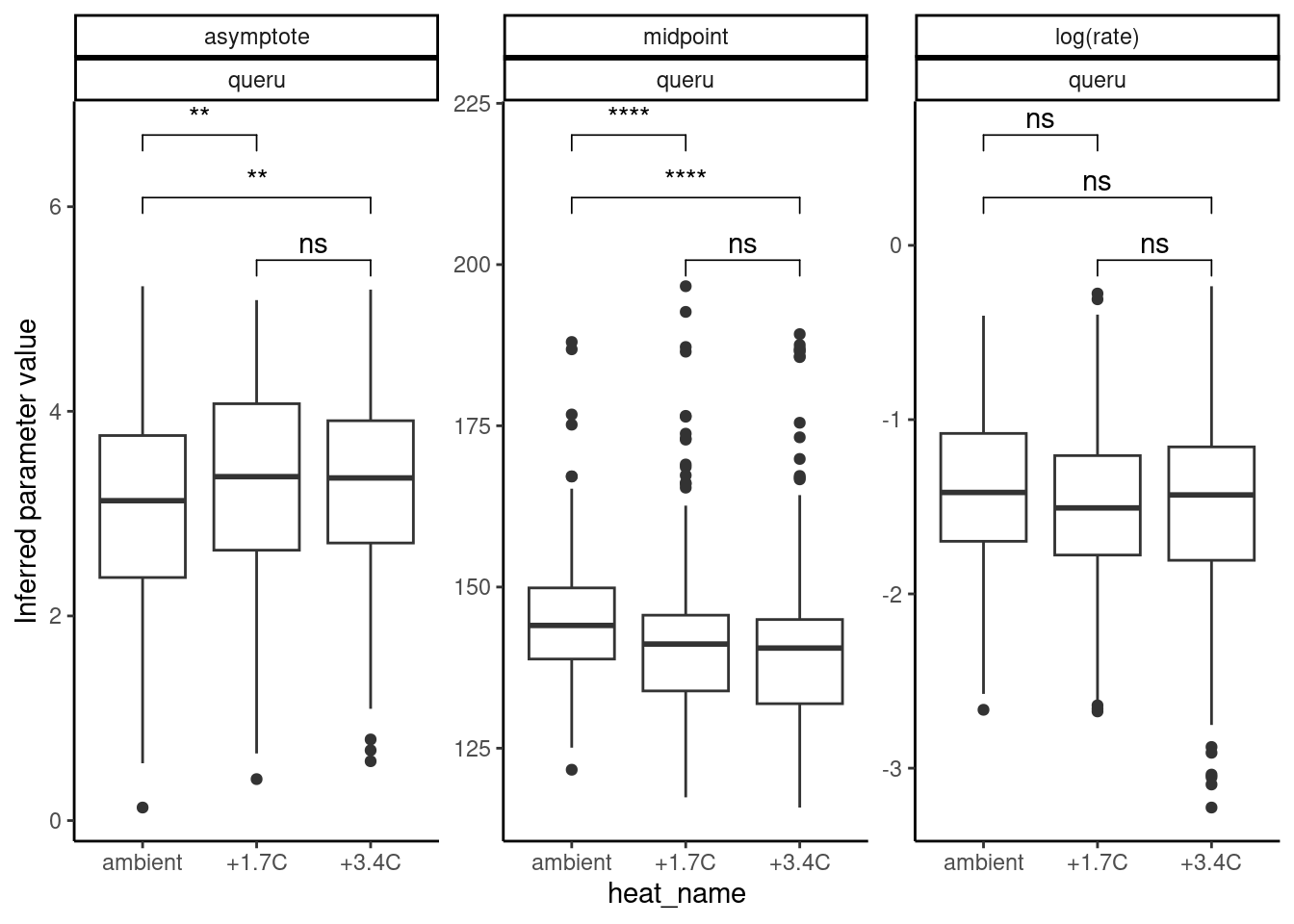
Infer treatment effects again.
plot_random_effects(df_mcmc = df_MCMC_12 %>% mutate(species = "queru"), dat = dat_12,
option = "relationship", var_x = "heat_name")
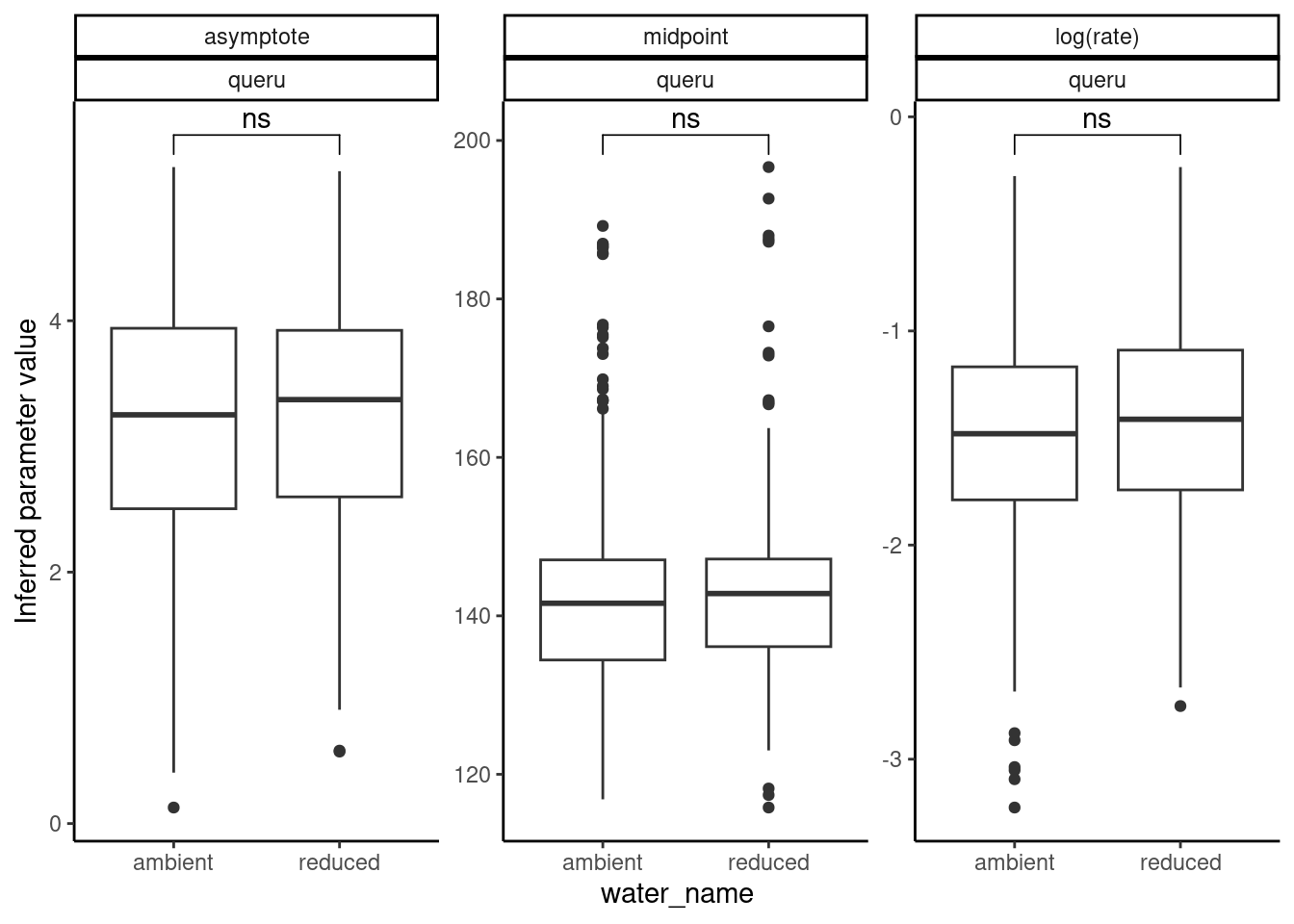
plot_random_effects(df_mcmc = df_MCMC_12 %>% mutate(species = "queru"), dat = dat_12,
option = "relationship", var_x = "water_name")
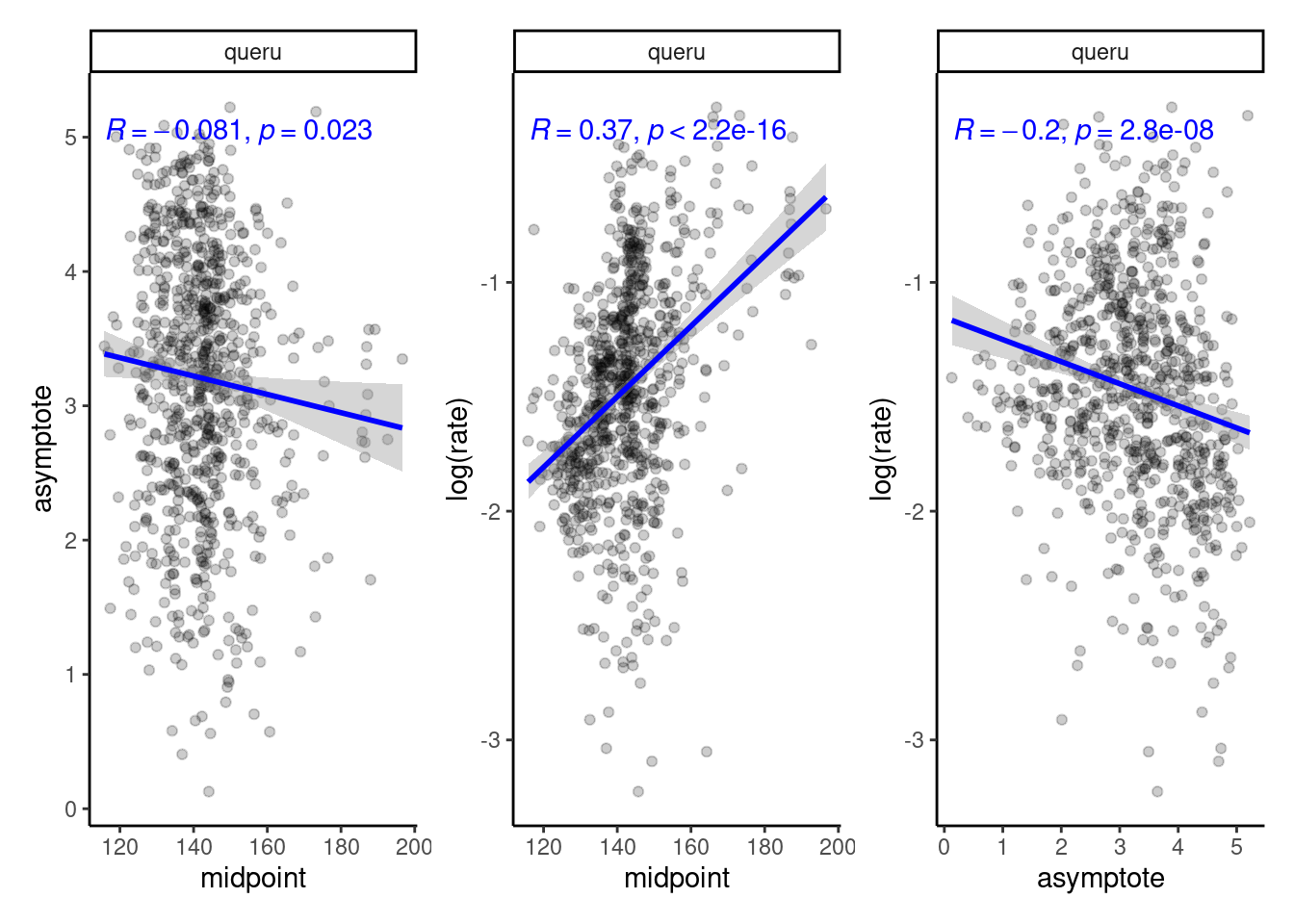
Any possible trade-off?
plot_random_effects(df_mcmc = df_MCMC_12 %>% mutate(species = "queru"), dat = dat_12,
option = "correlation")
Generalize to all species
dat_re <- dat_shoot_cov %>%
filter(shoot > 0) %>%
filter(doy > 90, doy <= 210) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = str_c(site, canopy, barcode, slot, year, sep = "_") %>% factor() %>% as.integer())
calc_bayes_re(
data = dat_re,
path = "alldata/intermediate/shootmodeling/re/",
num_cores = 10
)
plot_bayes_all (path = "alldata/intermediate/shootmodeling/re/")