Bayesian model 9
Based on Model 6. Site-year-level random effects. Ignore block-level, plot-level, individual-level differences. Treatments as covariates. Canopy as an additive term and interaction term with warming treatment.
Model
Data model
\begin{align*} y_{i,t,s,d} \sim \text{Lognormal}(\mu_{i,t,s,d}, \sigma^2) \end{align*}
Process model
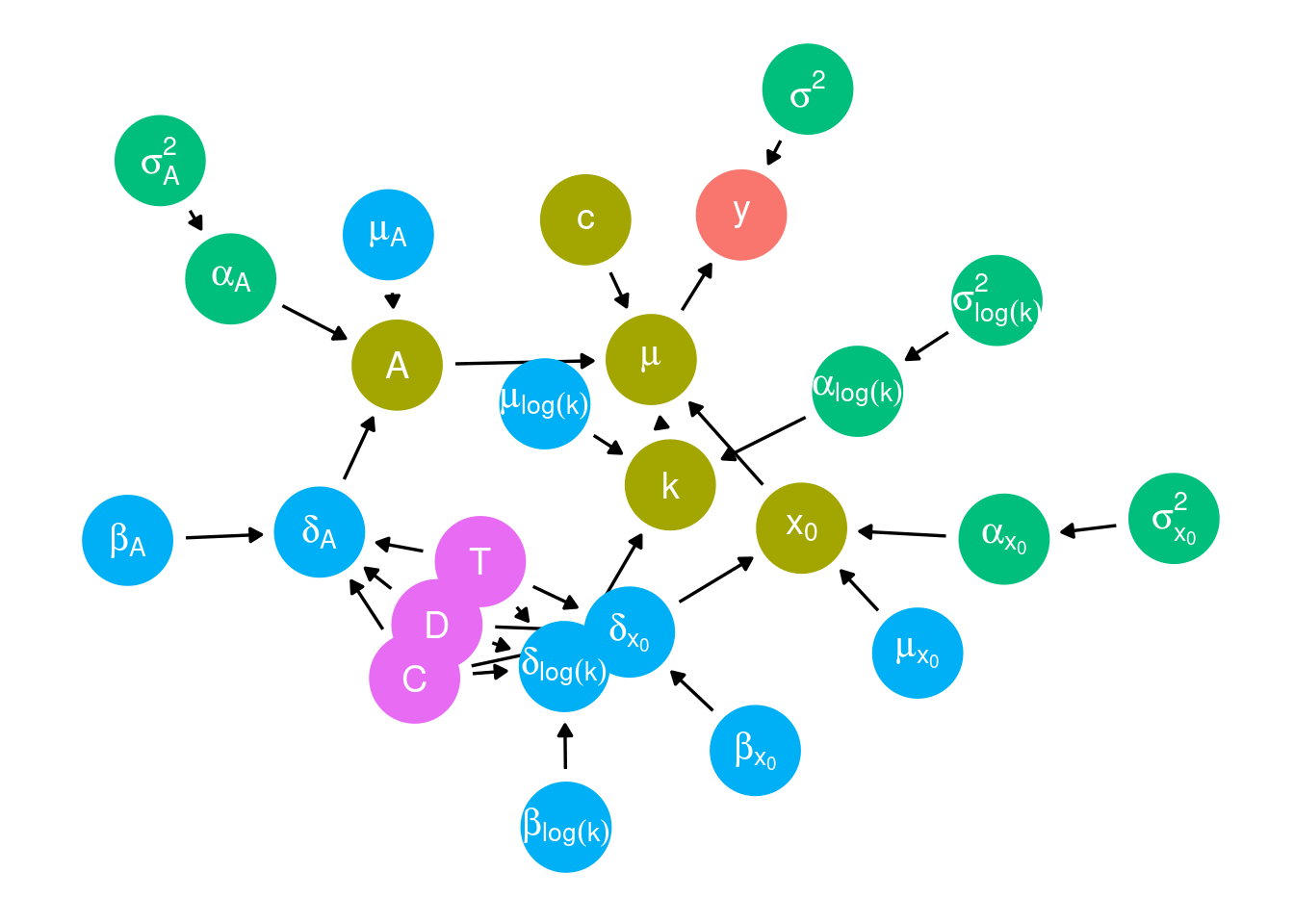
\begin{align*} \mu_{i,t,s,d} &= c+\frac{A_{i,t,s}}{1+e^{-k_{i,t,s}(d-x_{0 i,t,s})}} \newline A_{i,t,s} &= \mu_A + \delta_{A,i}+ \alpha_{A,t,s} \newline x_{0 i,t,s} &= \mu_{x_0} + \delta_{x_0,i}+ \alpha_{x_0,t,s} \newline log(k_{i,t,s}) &= \mu_{log(k)} + \delta_{log(k),i}+ \alpha_{log(k),t,s} \end{align*}
Fixed effects
\begin{align*} \delta_{A,i} &= \beta_{A,1} T_i + \beta_{A,2} D_i + \beta_{A,3} T_i D_i + \beta_{A,4} C_i + \beta_{A,5} T_i C_i \newline \delta_{x_0,i} &= \beta_{x_0,1} T_i + \beta_{x_0,2} D_i + \beta_{x_0,3} T_i D_i + \beta_{x_0,4} C_i + \beta_{x_0,5} T_i C_i \newline \delta_{log(k),i} &= \beta_{log(k),1} T_i + \beta_{log(k),2} D_i + \beta_{log(k),3} T_i D_i + \beta_{log(k),4} C_i + \beta_{log(k),5} T_i C_i \end{align*}
Random effects
\begin{align*} \alpha_{A,t,s} &\sim \text{Normal}(0, \sigma_A^2) \newline \alpha_{x_0,t,s} &\sim \text{Normal}(0, \sigma_{x_0}^2) \newline \alpha_{log(k),t,s} &\sim \text{Normal}(0, \sigma_{log(k)}^2) \end{align*}
Priors
\begin{align*} c &\sim \text{Normal}(0, 1) \newline \mu_A &\sim \text{Normal}(5, 1) \newline \beta_A &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 1 & 0 & 0 & 0 & 0 \newline 0 & 1 & 0 & 0 & 0 \newline 0 & 0 & 1 & 0 & 0 \newline 0 & 0 & 0 & 1 & 0 \newline 0 & 0 & 0 & 0 & 1 \newline \end{pmatrix} )\newline \sigma_A^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \mu_{x_0} &\sim \text{Normal}(160, 100) \newline \beta_{x_0} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 100 & 0 & 0 & 0 & 0 \newline 0 & 100 & 0 & 0 & 0 \newline 0 & 0 & 100 & 0 & 0 \newline 0 & 0 & 0 & 100 & 0 \newline 0 & 0 & 0 & 0 & 100 \newline \end{pmatrix} )\newline \sigma_{x_0}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \mu_{log(k)} &\sim \text{Normal}(-2, 0.04, 0, \infty) \newline \beta_{log(k)} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 0.04 & 0 & 0 & 0 & 0 \newline 0 & 0.04 & 0 & 0 & 0 \newline 0 & 0 & 0.04 & 0 & 0 \newline 0 & 0 & 0 & 0.04 & 0 \newline 0 & 0 & 0 & 0 & 0.04 \newline \end{pmatrix} )\newline \sigma_{log(k)}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \end{align*}
Draw a DAG.
plot_bayes_dag(version = 9)
Prepare data
dat_9 <- dat_shoot_cov %>%
filter(species == "queru") %>%
filter(doy > 90, doy <= 210) %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
mutate(group = str_c(site, year, sep = "_") %>% factor() %>% as.integer()) %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer()) %>%
mutate(canopy_code = factor(canopy, levels = c("open", "closed"), labels = c(0, 1)) %>% as.character() %>% as.integer())
Note that I selected for data between day 91 (Apr) and day 210 (Jul).
Fit model
df_MCMC_9 <- calc_bayes_fit(
data = dat_9 %>% mutate(tag = "training"),
version = 9,
num_iterations = 50000,
nthin = 5
)
write_rds(df_MCMC_9, "alldata/intermediate/shootmodeling/df_MCMC_9.rds")
df_MCMC_9 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_9.rds") %>%
tidy_mcmc(dat_9)
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_9)
p_bayes_diagnostics$p_MCMC
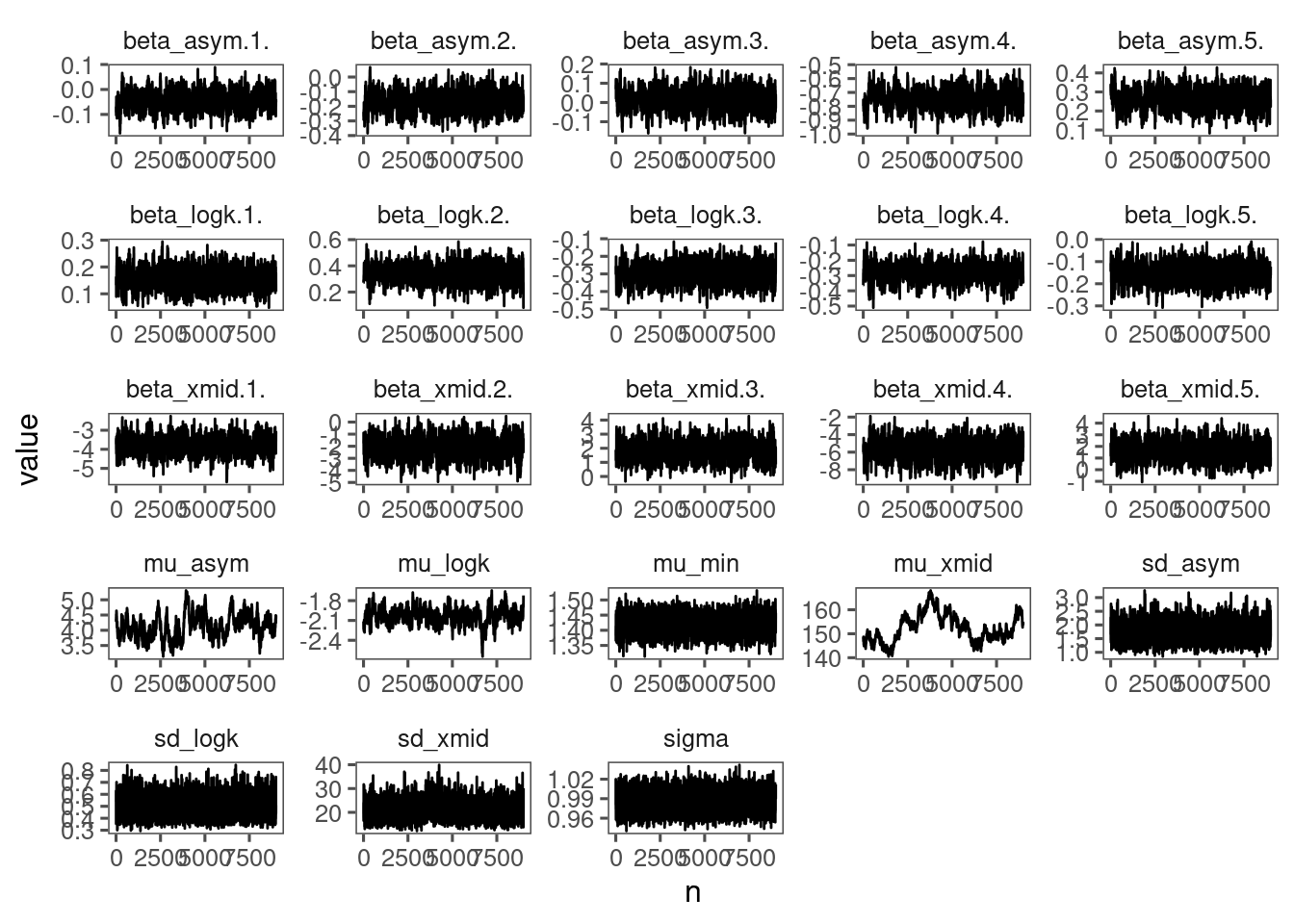 Speed is acceptable (~ 20 min). Mixing is ok. Using a longer chain (50,000 instead of 10,000 and thinned by a factor of 5).
Speed is acceptable (~ 20 min). Mixing is ok. Using a longer chain (50,000 instead of 10,000 and thinned by a factor of 5).
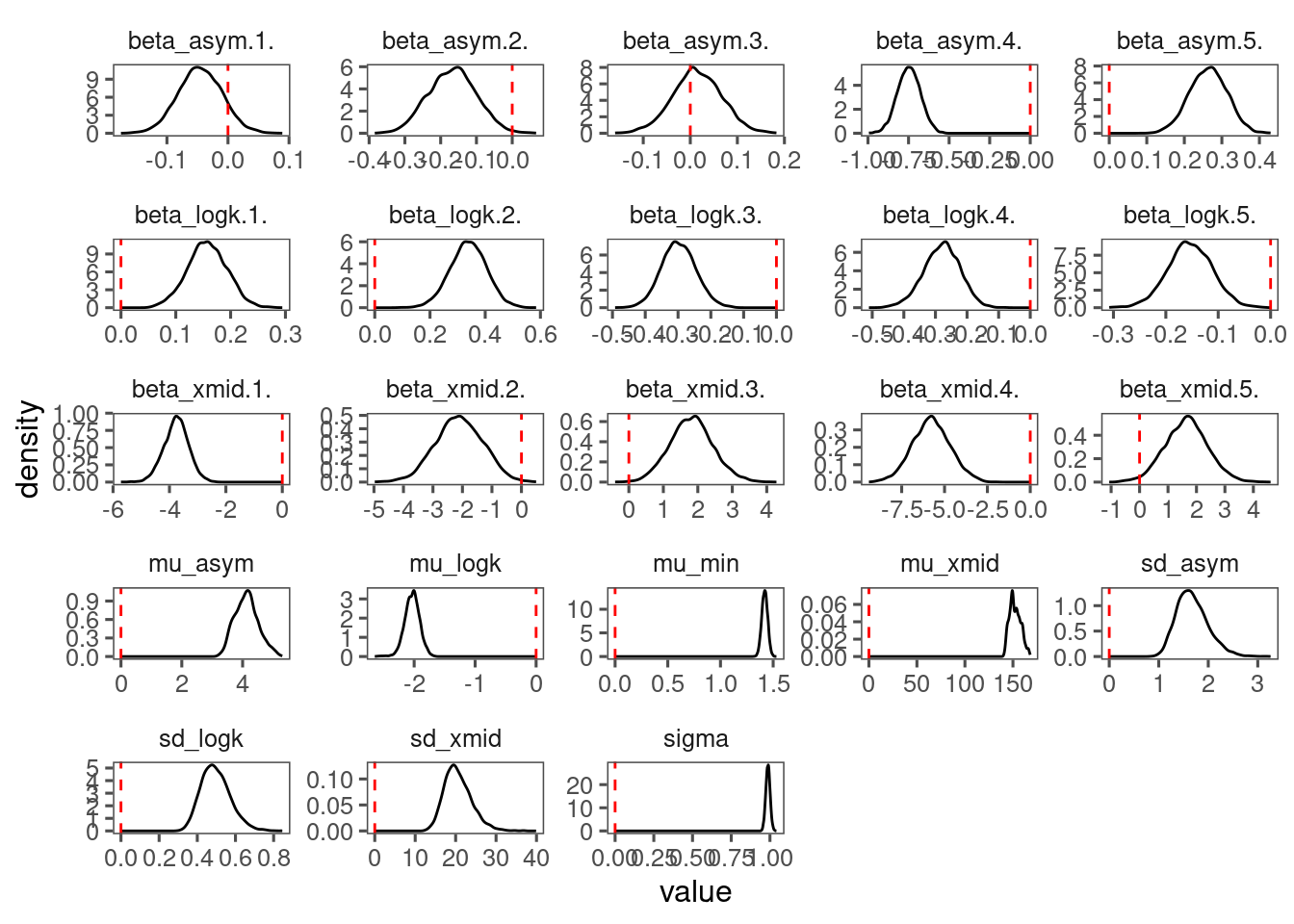
p_bayes_diagnostics$p_posterior
p_bayes_diagnostics$p_coefficient
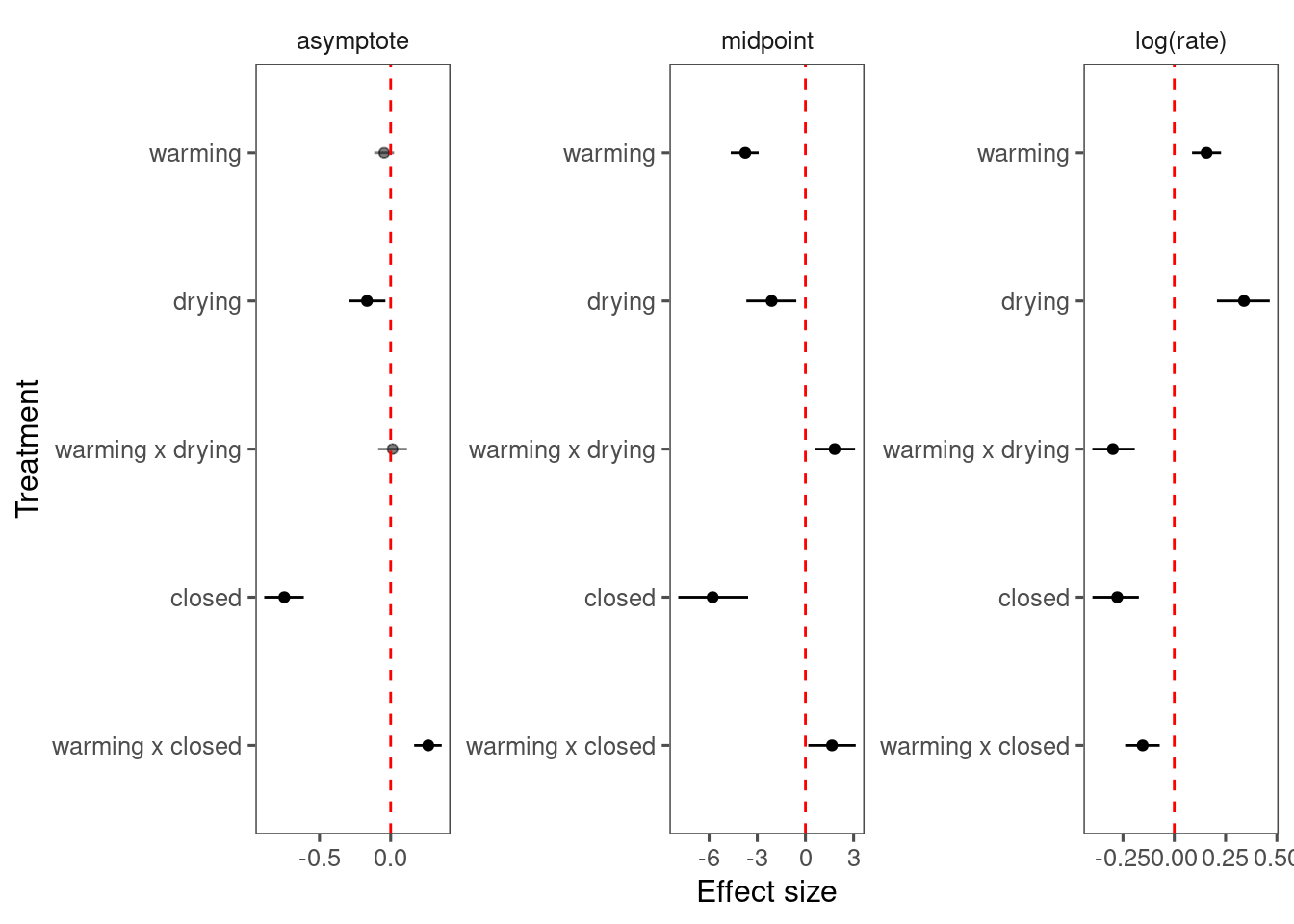
Inference:
- Drying and closed canopy reduce asymptote.
- Warming increases asymptote under closed canopy.
- Warming, drying, closed canopy advance midpoint.
- The advancing effect of warming is attenuated by drying and closed canopy condition.
- No significant effect on growth time
p_bayes_diagnostics$p_correlation
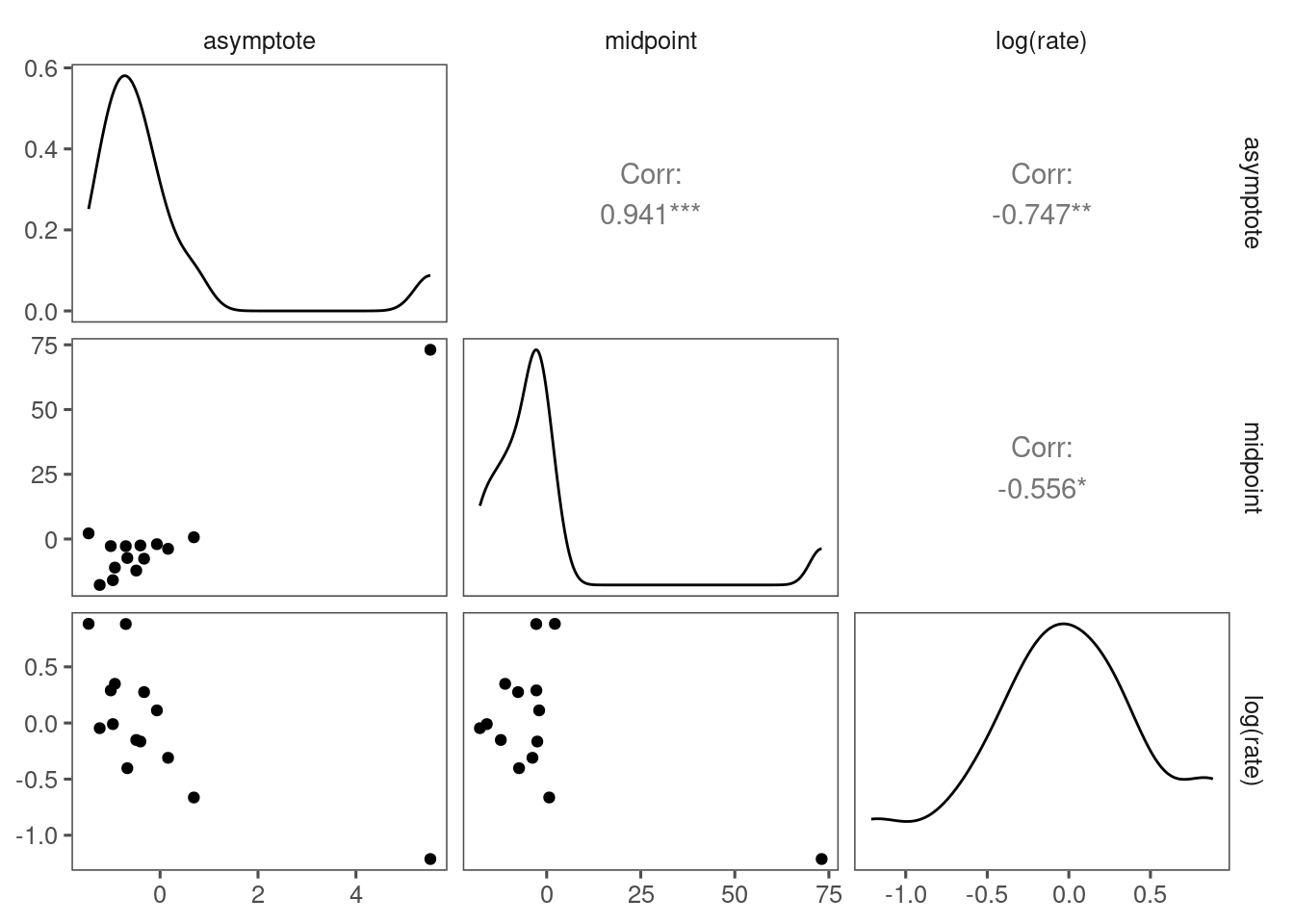 Did not see significant correlation among inferred random effects.
Did not see significant correlation among inferred random effects.
Make predictions
df_pred_9 <- calc_bayes_predict(
data = dat_9 %>%
distinct(site, year, group, heat_trt, water_trt, canopy, canopy_code, doy),
df_MCMC = df_MCMC_9,
version = 9
)
write_rds(df_pred_9, "alldata/intermediate/shootmodeling/df_pred_9.rds")
df_pred_9 <- read_rds("alldata/intermediate/shootmodeling/df_pred_9.rds")
p_bayes_predict <- plot_bayes_predict(
data = dat_9,
data_predict = df_pred_9,
vis_log = T
)
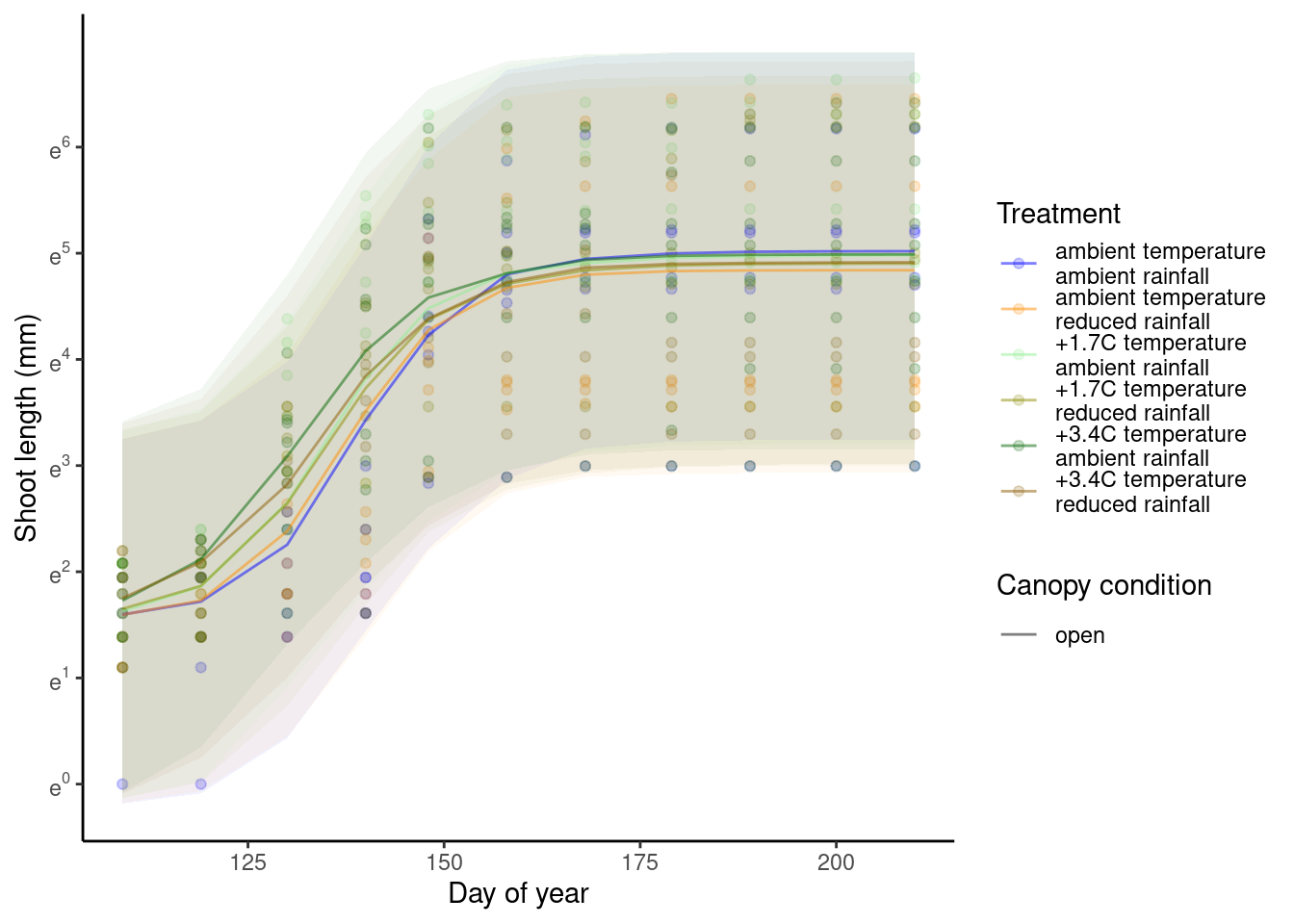
p_bayes_predict$p_original
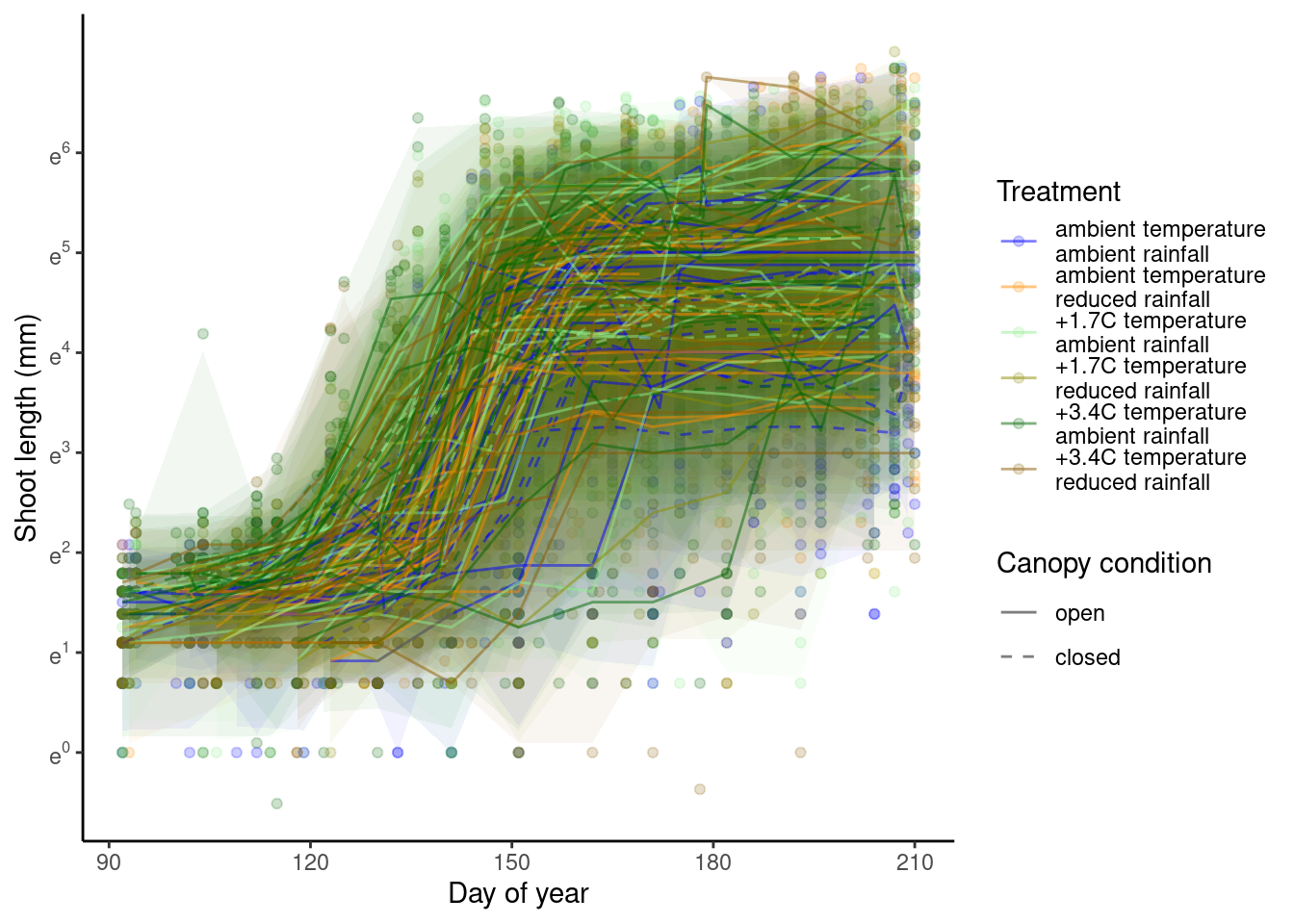
p_bayes_predict$p_overlay
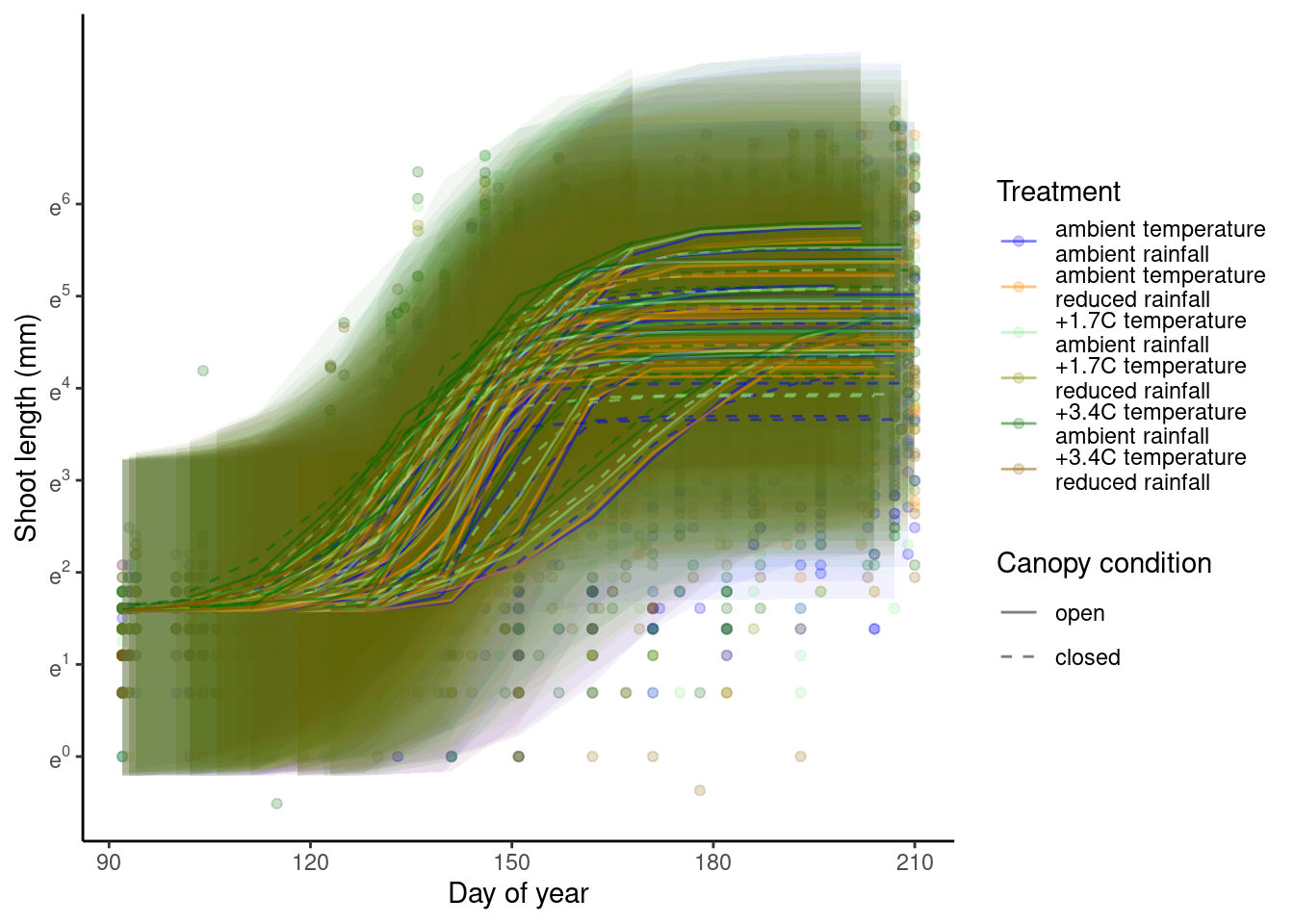
p_bayes_predict$p_accuracy
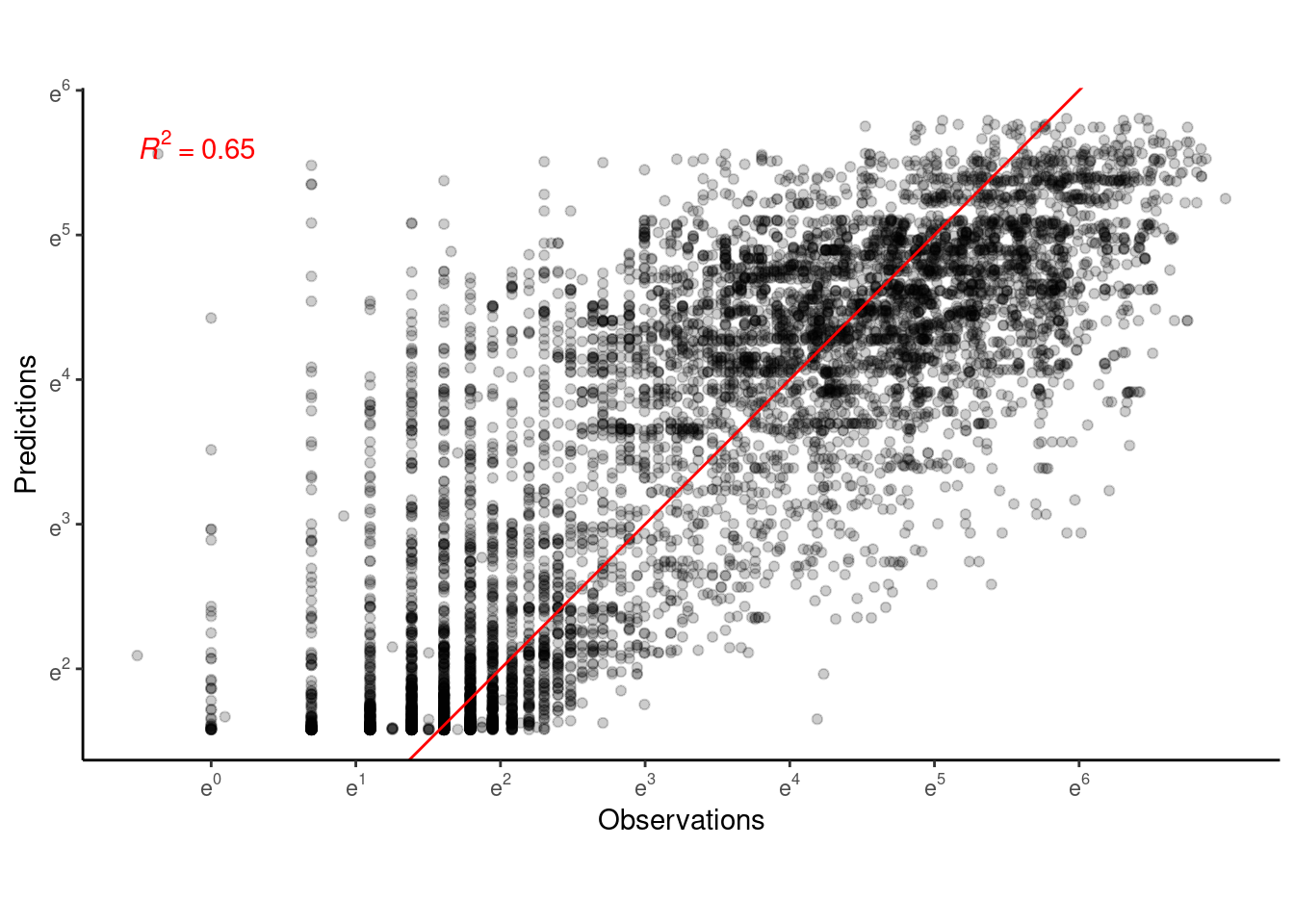
Visualize year 2016 and site HWRC alone.
p_bayes_predict <- plot_bayes_predict(
data = dat_9 %>% filter(year == 2016, site == "hwrc"),
data_predict = df_pred_9 %>% filter(year == 2016, site == "hwrc"),
vis_log = T
)
p_bayes_predict$p_original
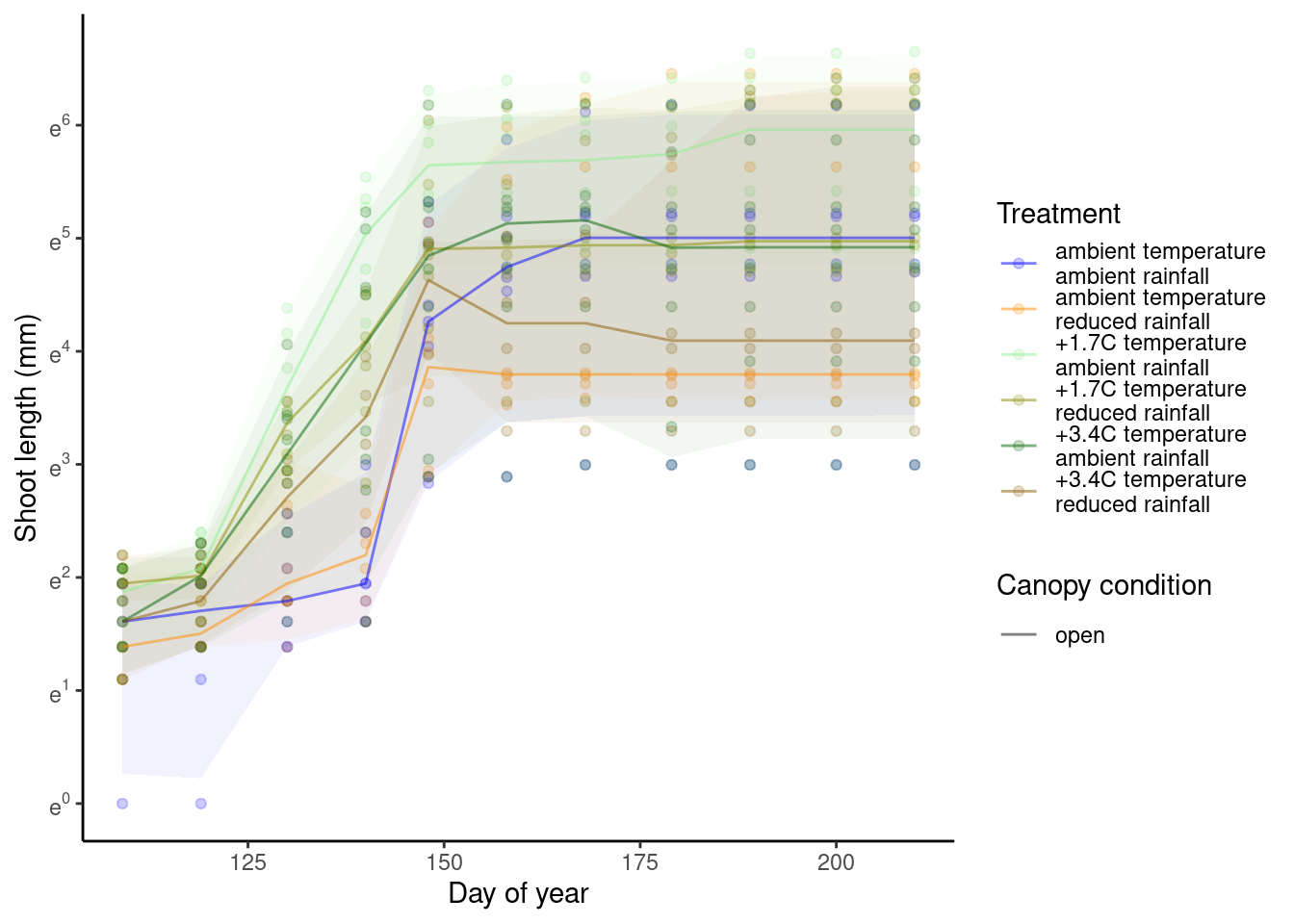
p_bayes_predict$p_overlay
Generalize to all species
Fit a separate model for each species.
Adjustments in models:
- For species with both canopy conditions and both water treatments, use Model 9.
- For species with one canopy condition and both water treatments, use Model 10.
- For species with one canopy condition and one water treatment, use Model 11.
dat_all <- dat_shoot_cov %>%
filter(doy > 90, doy <= 210) %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
group_by(species) %>%
mutate(group = str_c(site, year, sep = "_") %>% factor() %>% as.integer()) %>%
ungroup() %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer()) %>%
mutate(canopy_code = factor(canopy, levels = c("open", "closed"), labels = c(0, 1)) %>% as.character() %>% as.integer())
calc_bayes_all(
data = dat_all,
num_iterations = 50000,
nthin = 5,
path = "alldata/intermediate/shootmodeling/all/",
num_cores = 20
)
plot_bayes_all(path = "alldata/intermediate/shootmodeling/all/")
Here I only summarize the results from species with full factorial design (2 canopy conditions, 2 water treatments).
df_bayes_all <- read_bayes_all(path = "alldata/intermediate/shootmodeling/all/", full_factorial = T)
df_coef_summ <- summ_mcmc(df_bayes_all)
df_coef_summ %>%
arrange(response, covariate, desc(median)) %>%
select(response, covariate, category, median, lower, upper, species) %>%
group_by(response, covariate, category) %>%
summarise(species = toString(species), .groups = "drop")
## # A tibble: 54 × 4
## response covariate category species
## <fct> <fct> <fct> <chr>
## 1 asymptote warming positive (significant) acesa, betpa, quema
## 2 asymptote warming positive (non-significant) poptr, rhaca
## 3 asymptote warming negative (non-significant) queru, aceru
## 4 asymptote warming negative (significant) pinst, pinba, abiba, p…
## 5 asymptote drying positive (significant) rhaca
## 6 asymptote drying positive (non-significant) picgl
## 7 asymptote drying negative (non-significant) quema, poptr, acesa, a…
## 8 asymptote drying negative (significant) queru, aceru, betpa
## 9 asymptote warming x drying positive (significant) betpa, pinba
## 10 asymptote warming x drying positive (non-significant) poptr, aceru, rhaca, p…
## # ℹ 44 more rows
p_bayes_summ <- plot_bayes_summary(df_bayes_all)
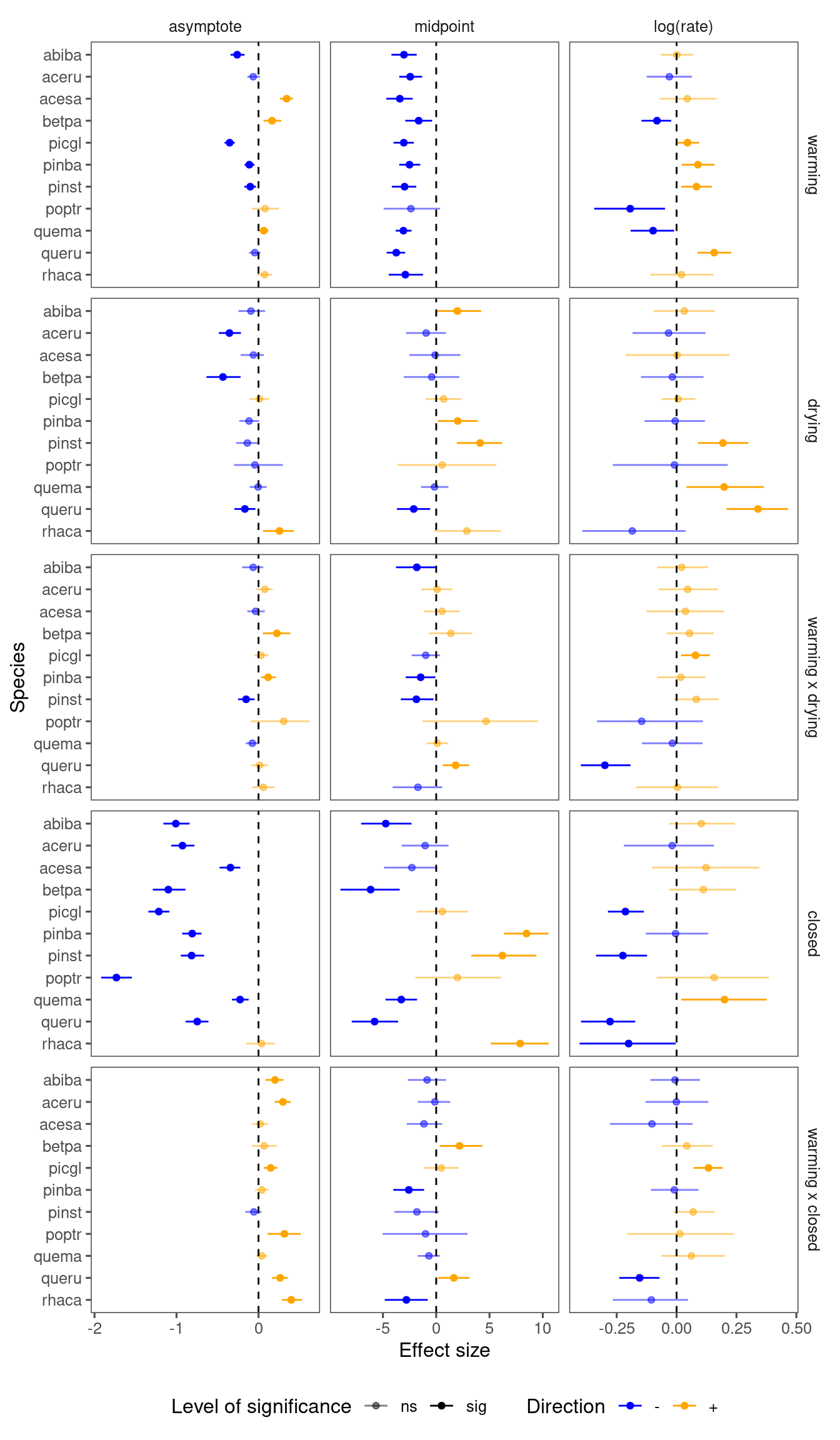
p_bayes_summ$p_coef_line
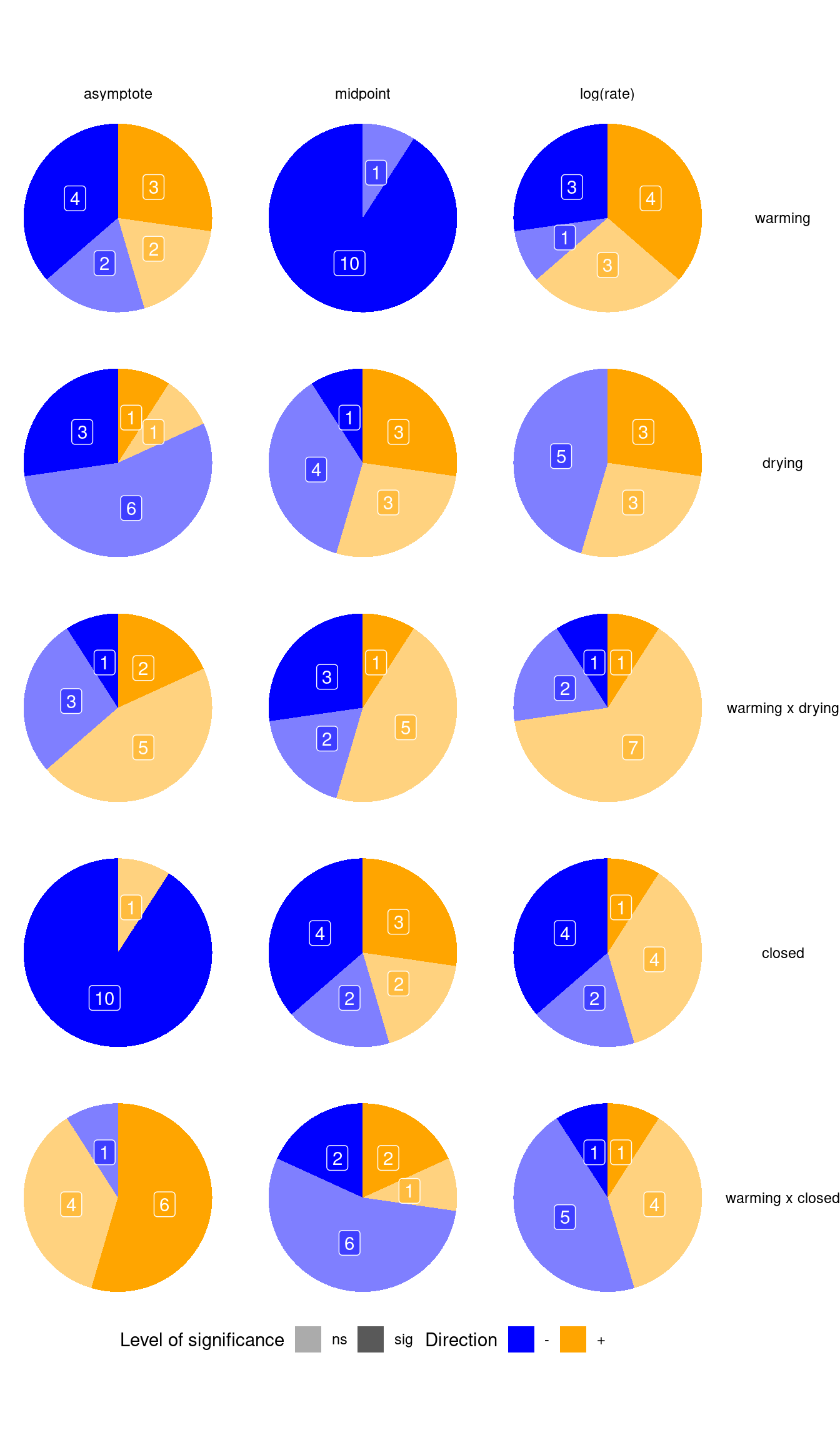
p_bayes_summ$p_coef_pie
p_bayes_summ$p_coef_pca
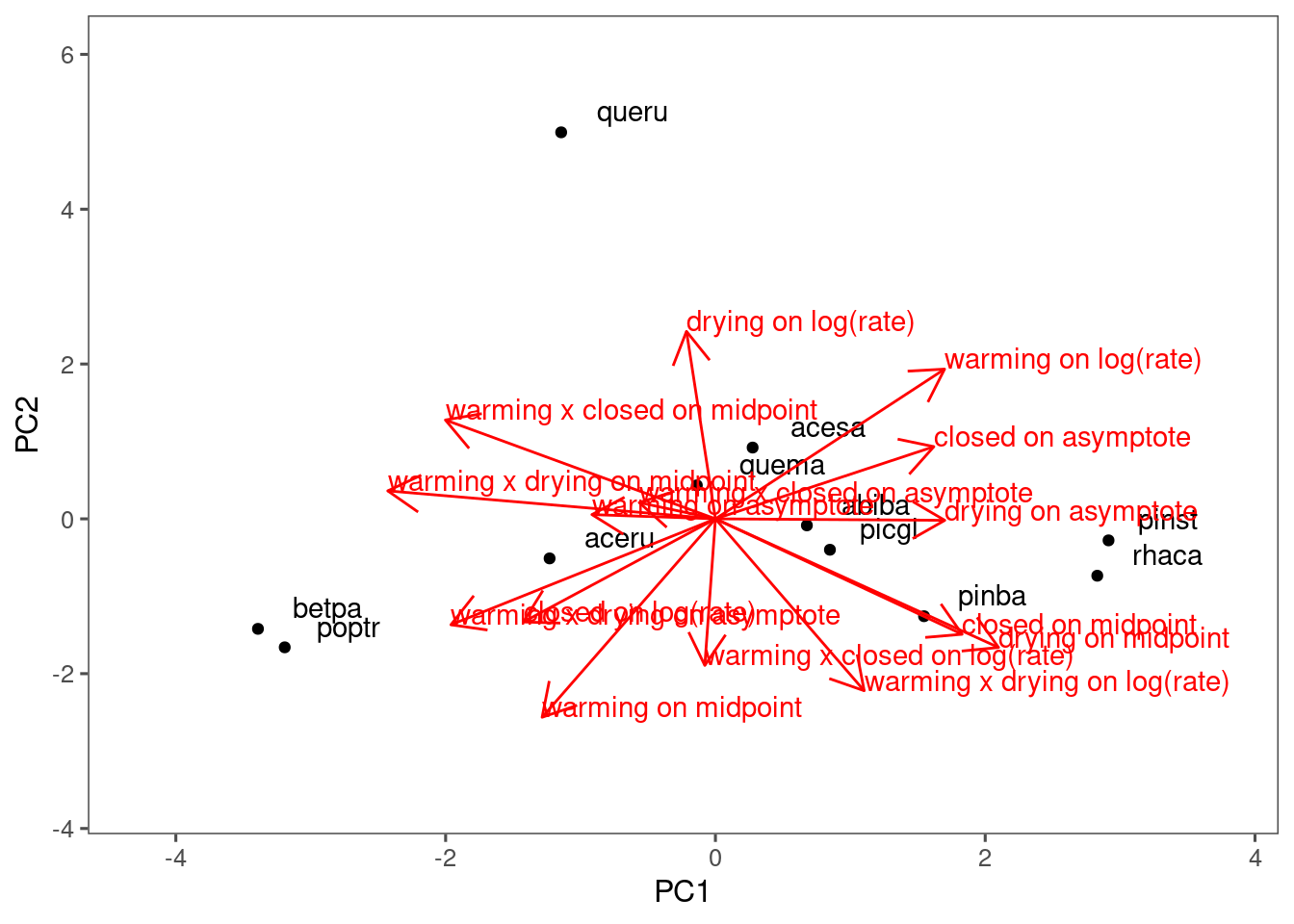
Climate change effects (summarizing the majority):
- Warming advances midpoint.
- Drying reduces asymptote, delays midpoint.
- Under drying, the effect of warming is more likely to further increase asymptote.
- Closed canopy reduces asymptote.
- Under closed canopy, the effect of warming is more likely to increase asymptote.
p_bayes_summ$p_corr_pie
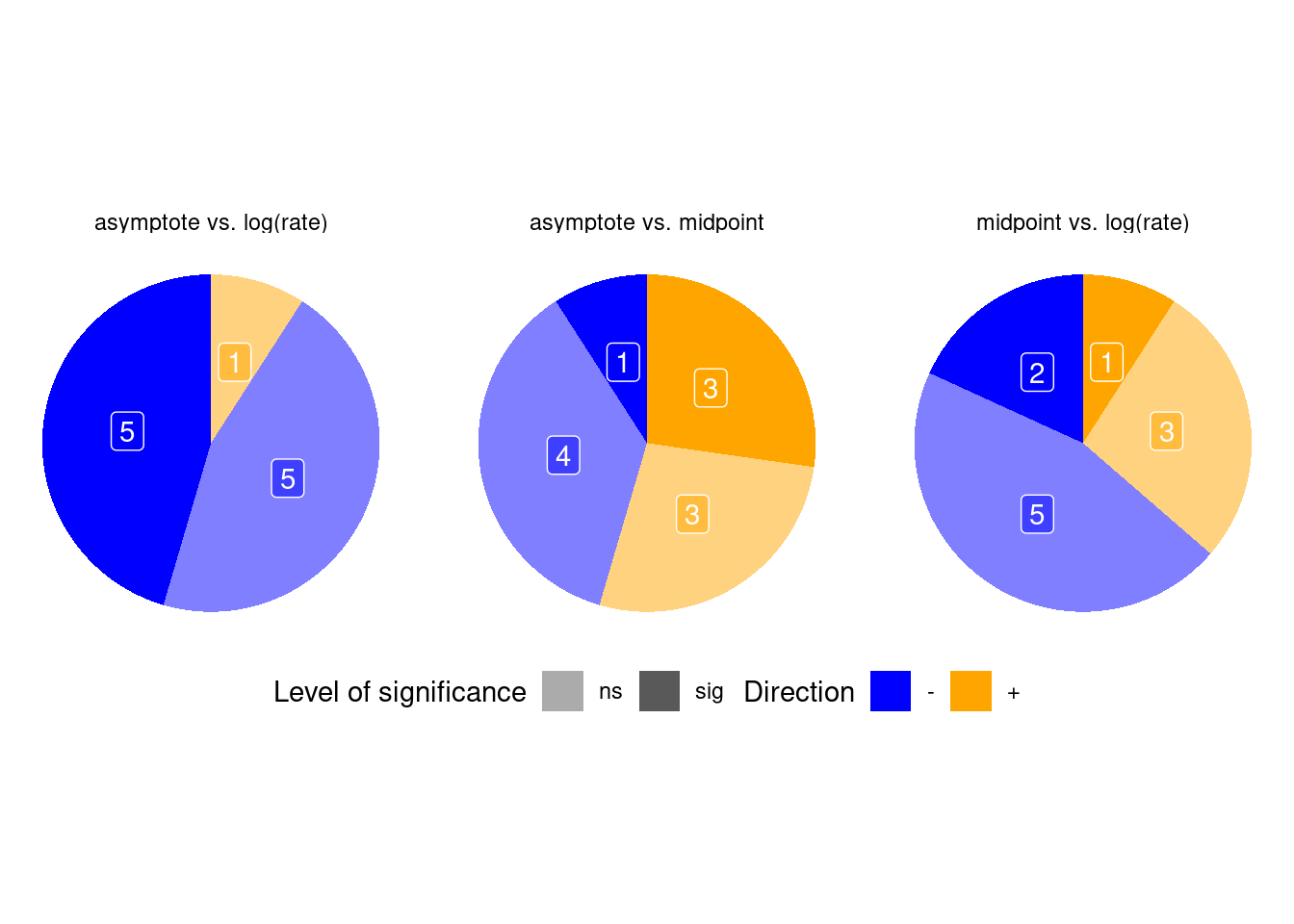
Correlation between parameters (summarizing the majority):
- Higher asympotote - longer growth time
- Higher asympotote - earlier midpoint
- Earlier midpoint - longer growth time