Bayesian model 5
Shoot-level random effects. Ignore year-level differences. Treatments as covariates.
Model
Data model
\begin{align*} y_{i,d} \sim \text{Lognormal}(\mu_{i,d}, \sigma^2) \end{align*}
Process model
\begin{align*}
\mu_{i,d} &= c+\frac{A_{i}}{1+e^{-k_{i}(d-x_{0 i})}} \newline
A_{i} &= \mu_A + \delta_{A,i}+ \alpha_{A,i} \newline
x_{0 i} &= \mu_{x_0} + \delta_{x_0,i}+ \alpha_{x_0,i} \newline
log(k_{i}) &= \mu_{log(k)} + \delta_{log(k),i}+ \alpha_{log(k),i}
\end{align*}
Fixed effects
\begin{align*} \delta_{A,i} &= \beta_{A,1} T_i + \beta_{A,2} D_i + \beta_{A,3} T_i D_i \newline \delta_{x_0,i} &= \beta_{x_0,1} T_i + \beta_{x_0,2} D_i + \beta_{x_0,3} T_i D_i \newline \delta_{log(k),i} &= \beta_{log(k),1} T_i + \beta_{log(k),2} D_i + \beta_{log(k),3} T_i D_i \end{align*}
Random effects
\begin{align*} \alpha_{A,i} &\sim \text{Normal}(0, \sigma_A^2) \newline \alpha_{x_0,i} &\sim \text{Normal}(0, \sigma_{x_0}^2) \newline \alpha_{log(k),i} &\sim \text{Normal}(0, \sigma_{log(k)}^2) \end{align*}
Priors
\begin{align*} c &\sim \text{Normal}(0, 1) \newline \mu_A &\sim \text{Normal}(5, 1) \newline \beta_A &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 1 & 0 & 0 \newline 0 & 1 & 0 \newline 0 & 0 & 1 \newline \end{pmatrix} )\newline \sigma_A^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \mu_{x_0} &\sim \text{Normal}(160, 100) \newline \beta_{x_0} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 100 & 0 & 0 \newline 0 & 100 & 0 \newline 0 & 0 & 100 \newline \end{pmatrix} )\newline \sigma_{x_0}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \mu_{log(k)} &\sim \text{Normal}(-2, 0.04) \newline \beta_{log(k)} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 0.04 & 0 & 0 \newline 0 & 0.04 & 0 \newline 0 & 0 & 0.04 \newline \end{pmatrix} )\newline \sigma_{log(k)}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \end{align*}
Prepare data
dat_5 <- dat_shoot_cov %>%
filter(species == "queru", canopy == "open", site == "cfc") %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = str_c(barcode, slot, sep = "_") %>% factor() %>% as.integer()) %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer())
Fit model
df_MCMC_5 <- calc_bayes_fit(
data = dat_5 %>% mutate(tag = "training"),
version = 5
)
write_rds(df_MCMC_5, "alldata/intermediate/shootmodeling/df_MCMC_5.rds")
df_MCMC_5 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_5.rds") %>% tidy_mcmc(dat_5)
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_5, plot_corr = F)
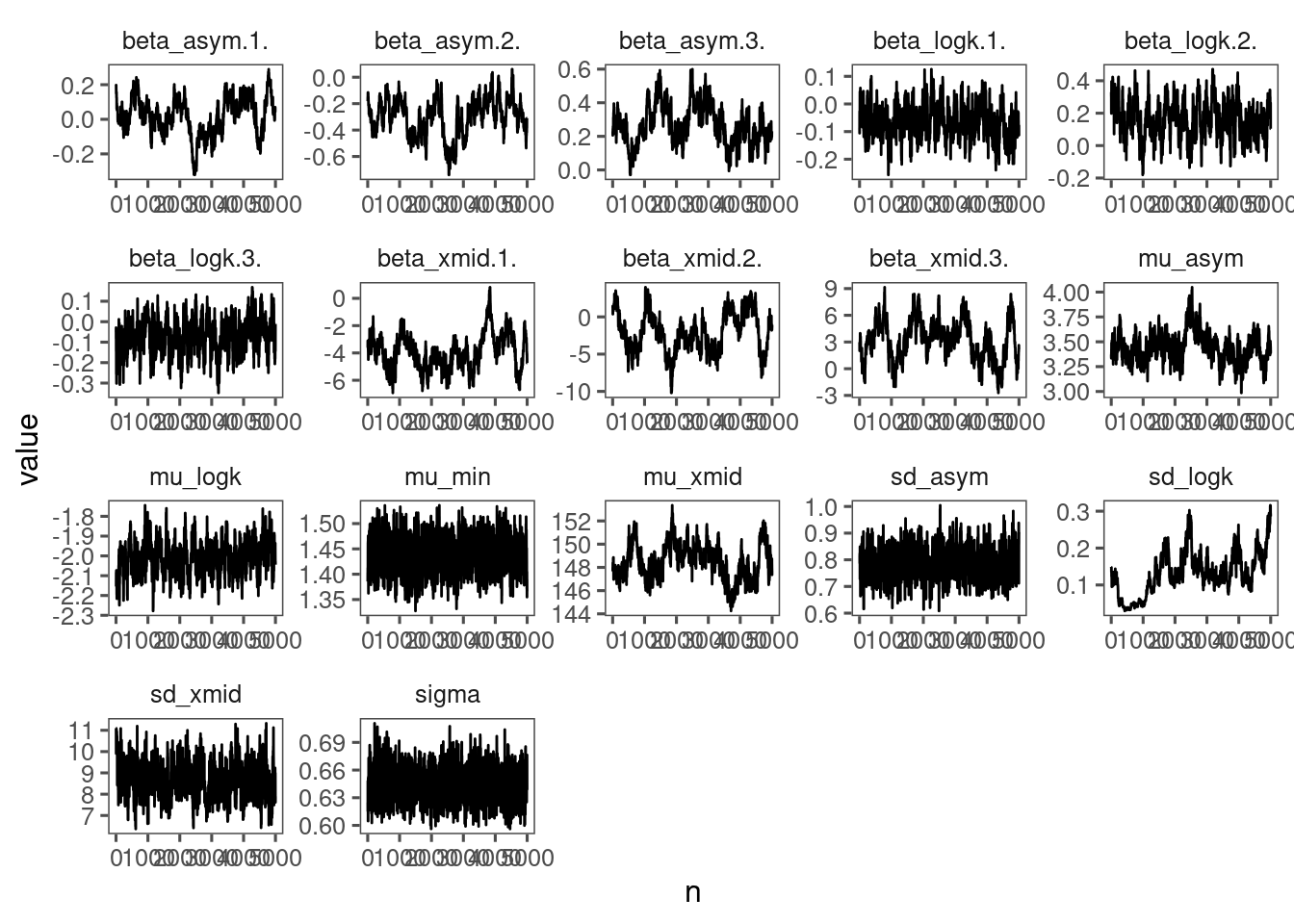
p_bayes_diagnostics$p_MCMC
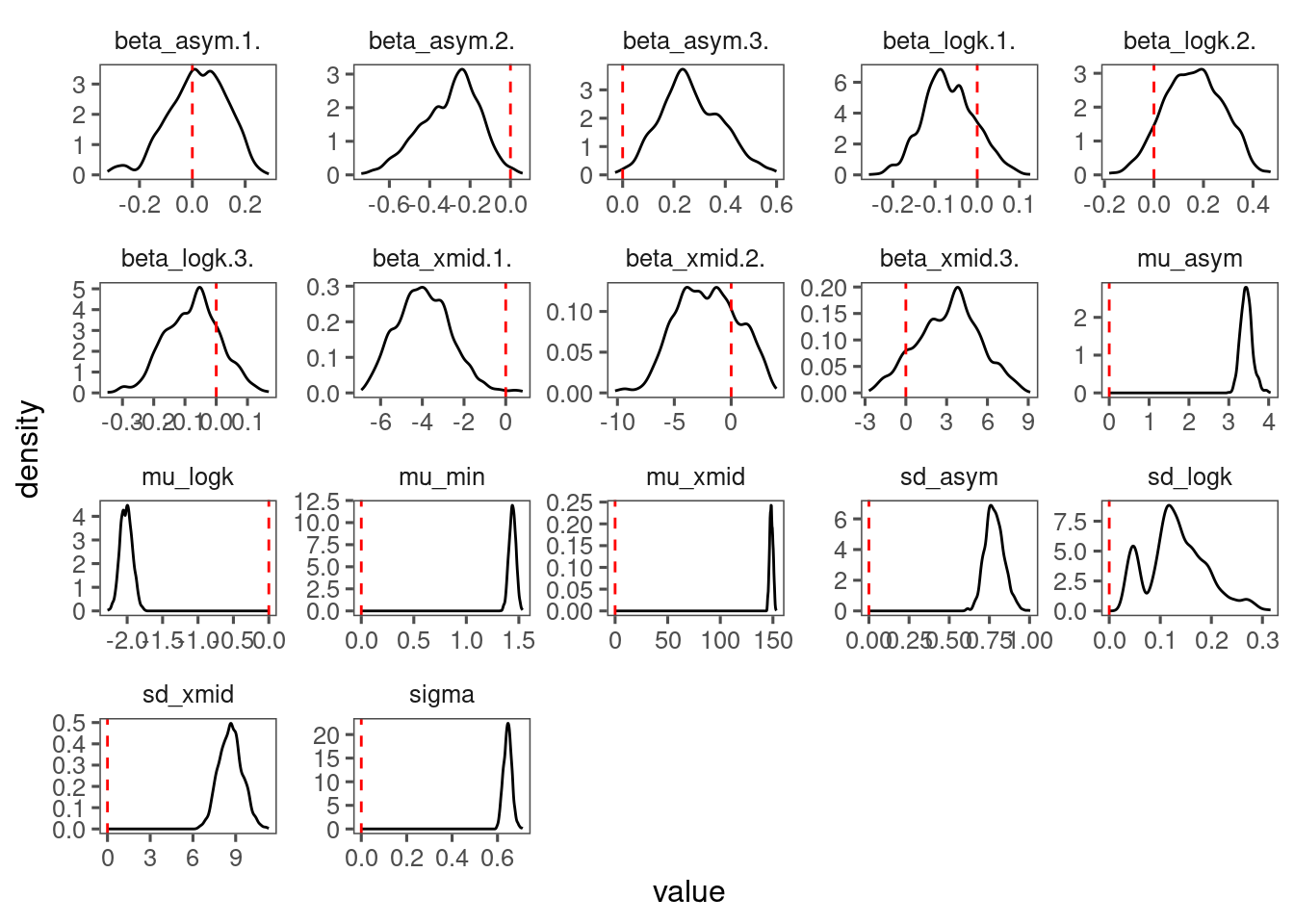
p_bayes_diagnostics$p_posterior
p_bayes_diagnostics$p_coefficient
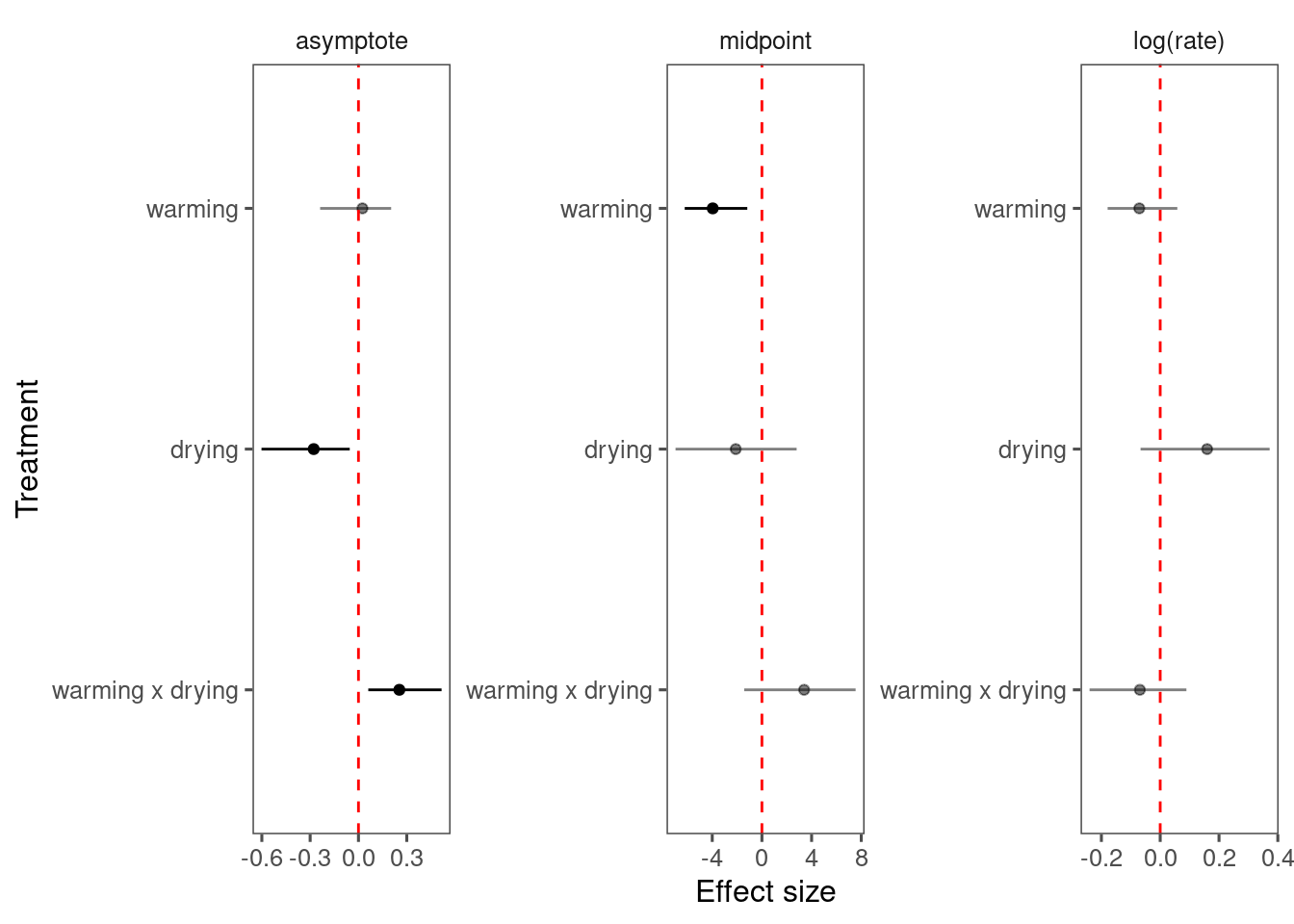
Mixing only slightly better than model 4 but still too many individuals. Might need longer MCMC chains.
Make predictions
df_pred_5 <- calc_bayes_predict(
data = dat_5 %>%
distinct(group, heat_trt, water_trt, doy),
df_MCMC = df_MCMC_5,
version = 5
)
write_rds(df_pred_5, "alldata/intermediate/shootmodeling/df_pred_5.rds")
df_pred_5 <- read_rds("alldata/intermediate/shootmodeling/df_pred_5.rds")
p_bayes_predict <- plot_bayes_predict(
data = dat_5,
data_predict = df_pred_5,
vis_log = T
)
p_bayes_predict$p_original
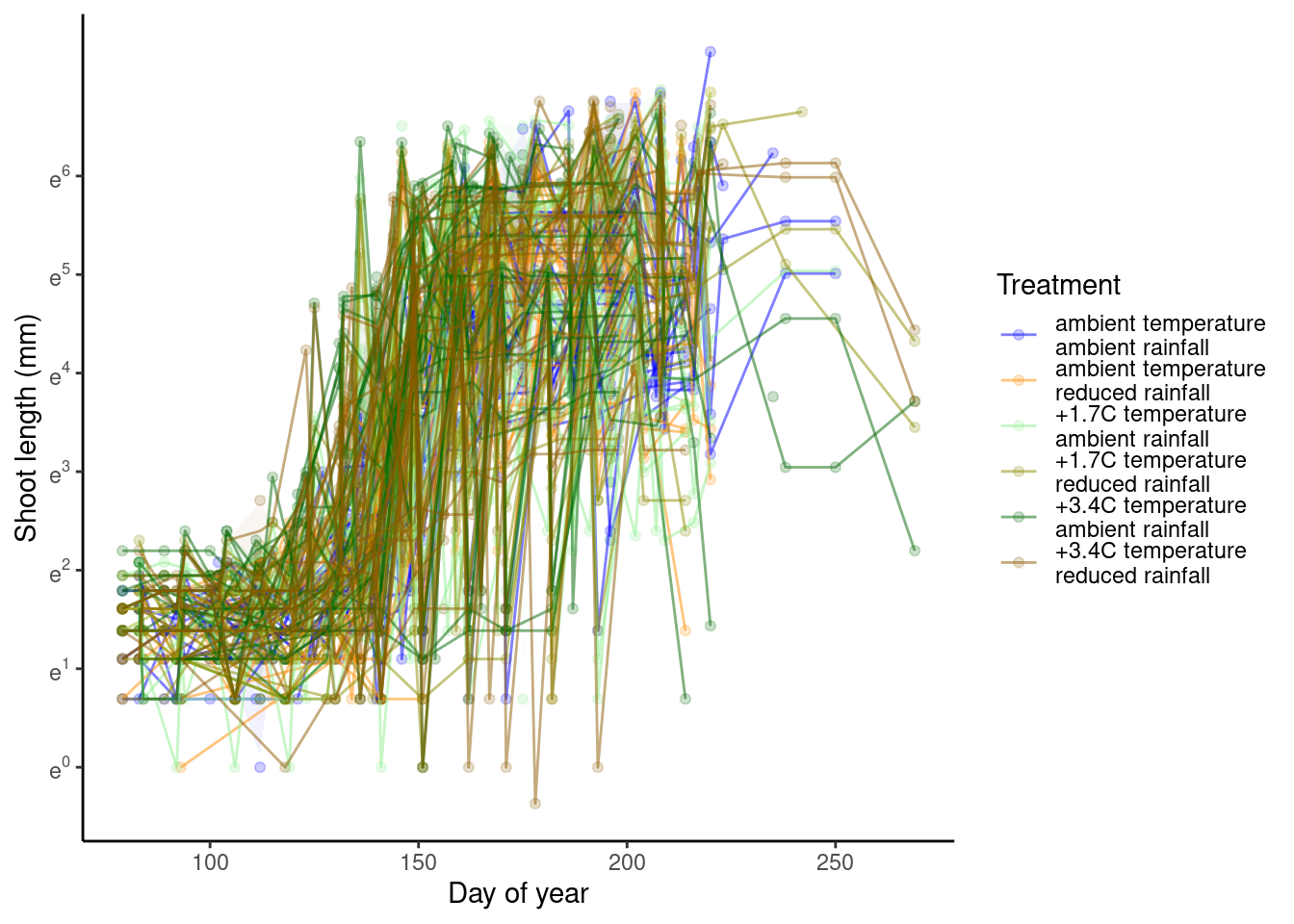
p_bayes_predict$p_overlay
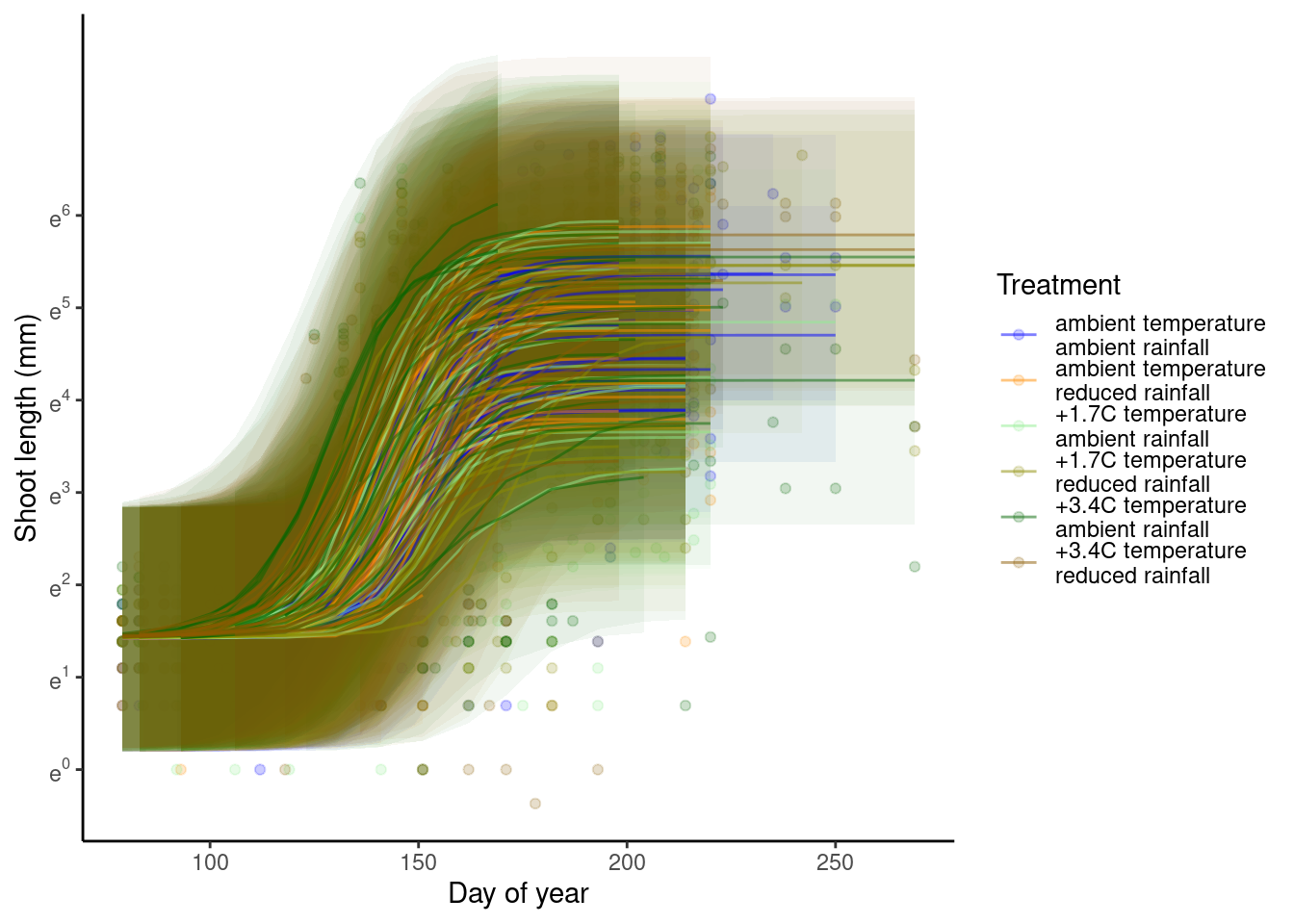
p_bayes_predict$p_accuracy
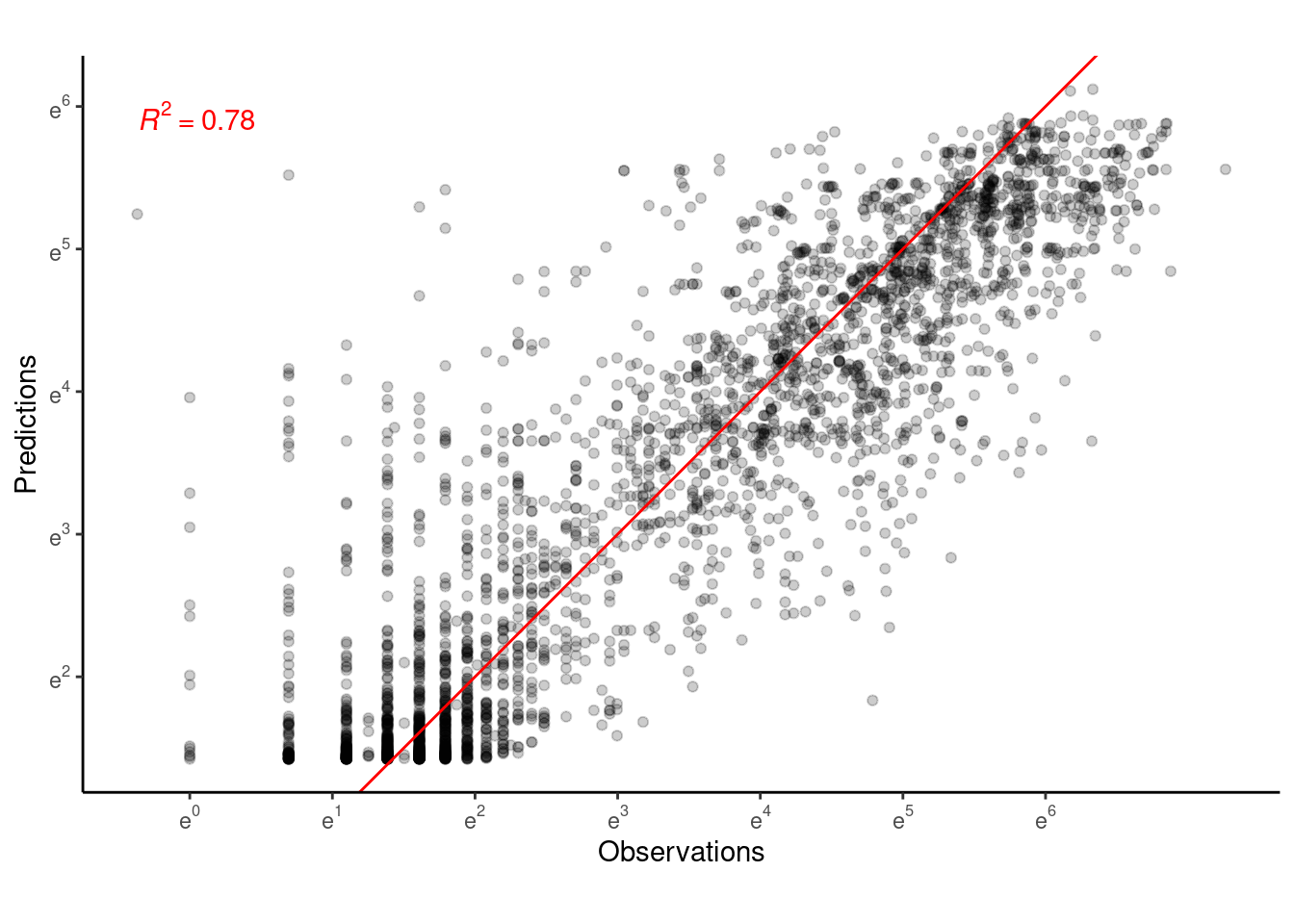
Bayesian model 6
Year-level random effects. Ignore shoot-level differences. Treatments as covariates.
Model
Data model
\begin{align*} y_{i,t,d} \sim \text{Lognormal}(\mu_{i,t,d}, \sigma^2) \end{align*}
Process model
\begin{align*} \mu_{i,t,d} &= c+\frac{A_{i,t}}{1+e^{-k_{i,t}(d-x_{0 i,t})}} \newline A_{i,t} &= \mu_A + \delta_{A,i}+ \alpha_{A,t} \newline x_{0 i,t} &= \mu_{x_0} + \delta_{x_0,i}+ \alpha_{x_0,t} \newline log(k_{i,t}) &= \mu_{log(k)} + \delta_{log(k),i}+ \alpha_{log(k),t} \end{align*}
Fixed effects
\begin{align*} \delta_{A,i} &= \beta_{A,1} T_i + \beta_{A,2} D_i + \beta_{A,3} T_i D_i \newline \delta_{x_0,i} &= \beta_{x_0,1} T_i + \beta_{x_0,2} D_i + \beta_{x_0,3} T_i D_i \newline \delta_{log(k),i} &= \beta_{log(k),1} T_i + \beta_{log(k),2} D_i + \beta_{log(k),3} T_i D_i \end{align*}
Random effects
\begin{align*} \alpha_{A,t} &\sim \text{Normal}(0, \sigma_A^2) \newline \alpha_{x_0,t} &\sim \text{Normal}(0, \sigma_{x_0}^2) \newline \alpha_{log(k),t} &\sim \text{Normal}(0, \sigma_{log(k)}^2) \end{align*}
Priors
c &\sim \text{Normal}(0, 1) \newline \begin{align*} \mu_A &\sim \text{Normal}(5, 1) \newline \beta_A &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 1 & 0 & 0 \newline 0 & 1 & 0 \newline 0 & 0 & 1 \newline \end{pmatrix} )\newline \sigma_A^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \mu_{x_0} &\sim \text{Normal}(160, 100) \newline \beta_{x_0} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 100 & 0 & 0 \newline 0 & 100 & 0 \newline 0 & 0 & 100 \newline \end{pmatrix} )\newline \sigma_{x_0}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \mu_{log(k)} &\sim \text{Normal}(-2, 0.04) \newline \beta_{log(k)} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 0.04 & 0 & 0 \newline 0 & 0.04 & 0 \newline 0 & 0 & 0.04 \newline \end{pmatrix} )\newline \sigma_{log(k)}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \end{align*}
Prepare data
dat_6 <- dat_shoot_cov %>%
filter(species == "queru", canopy == "open", site == "cfc") %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = year %>% factor() %>% as.integer()) %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer())
Fit model
df_MCMC_6 <- calc_bayes_fit(
data = dat_6 %>% mutate(tag = "training"),
version = 6
)
write_rds(df_MCMC_6, "alldata/intermediate/shootmodeling/df_MCMC_6.rds")
df_MCMC_6 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_6.rds") %>% tidy_mcmc(dat_6)
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_6, plot_corr = F)
p_bayes_diagnostics$p_MCMC
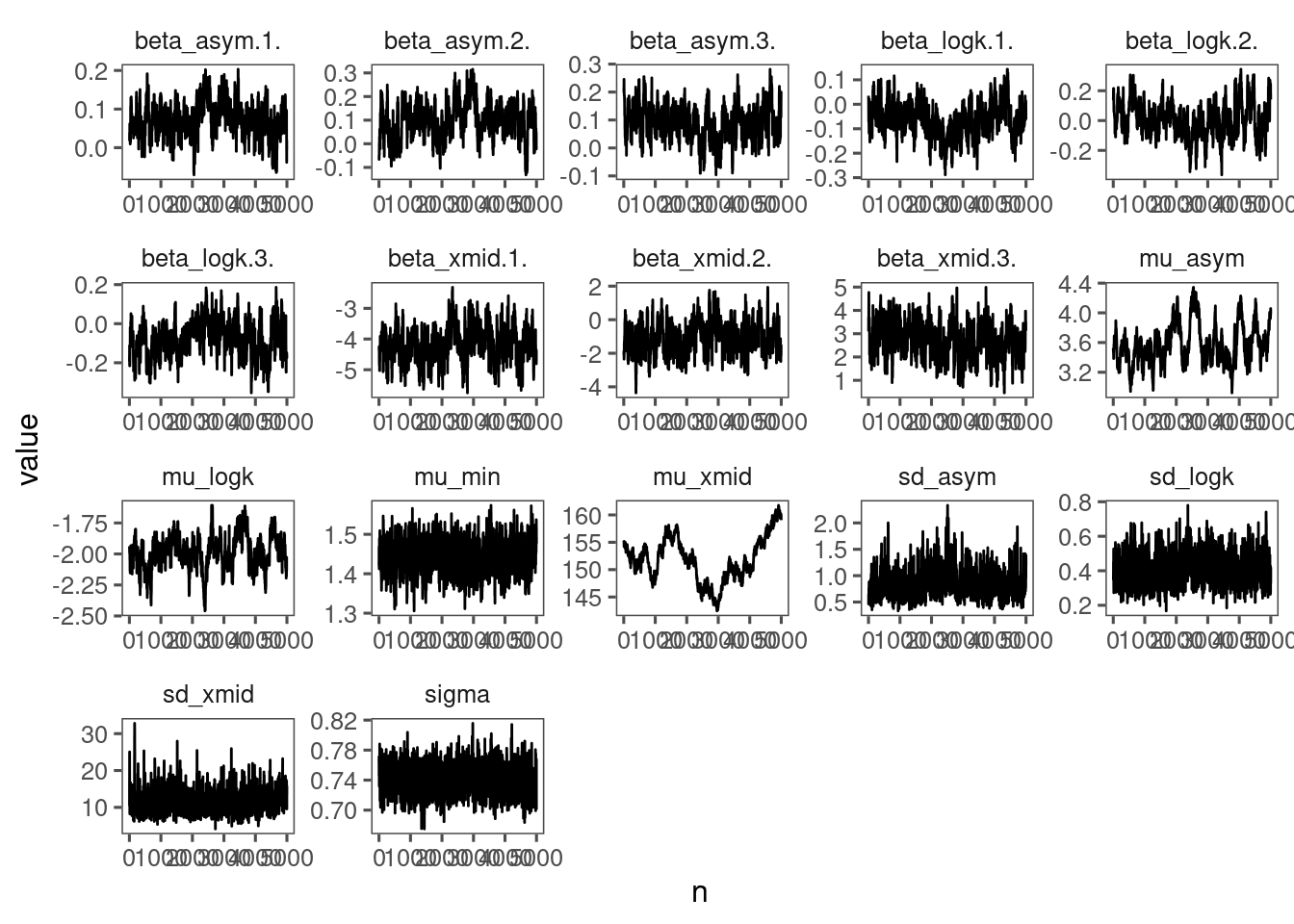
p_bayes_diagnostics$p_posterior
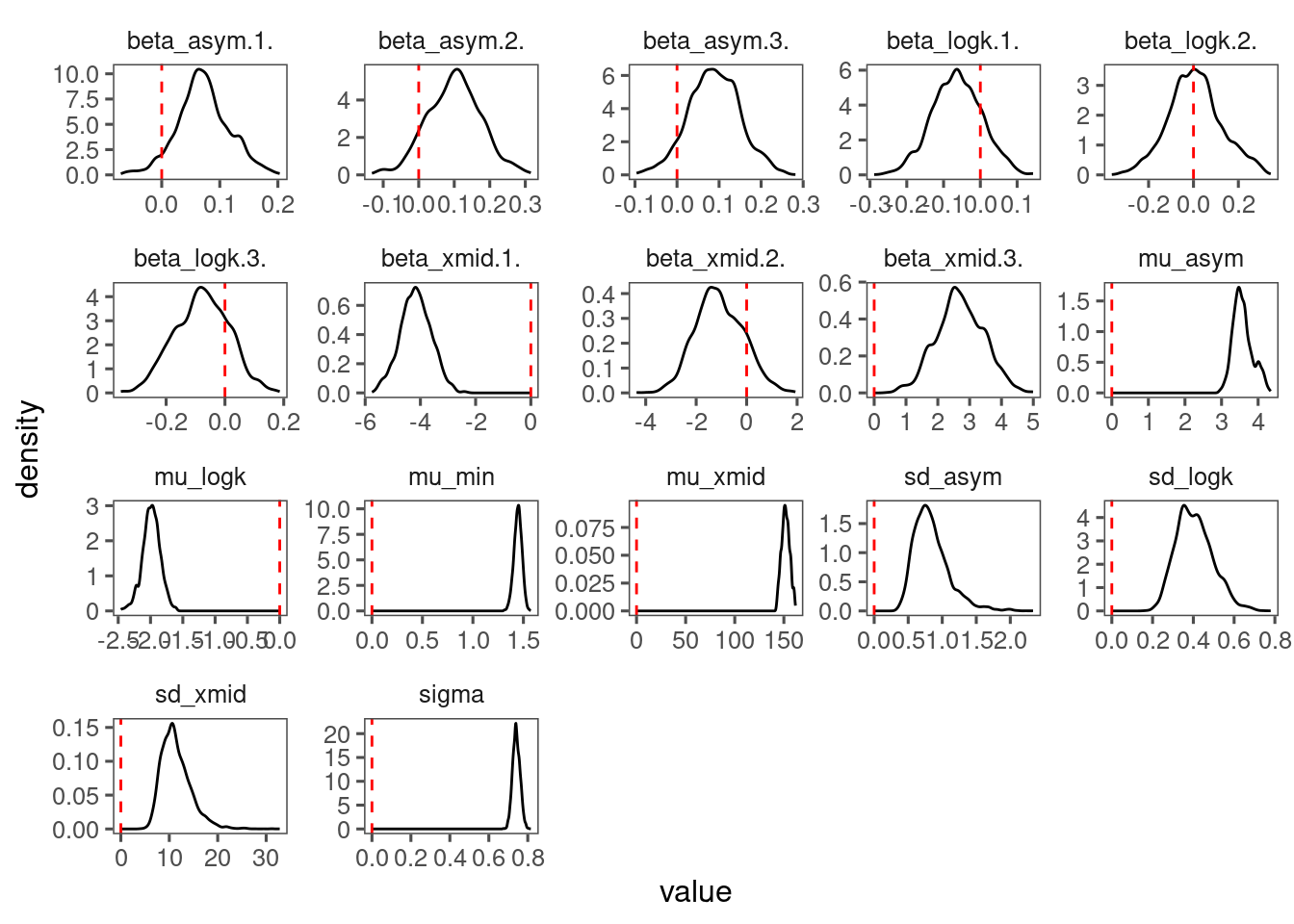
p_bayes_diagnostics$p_coefficient
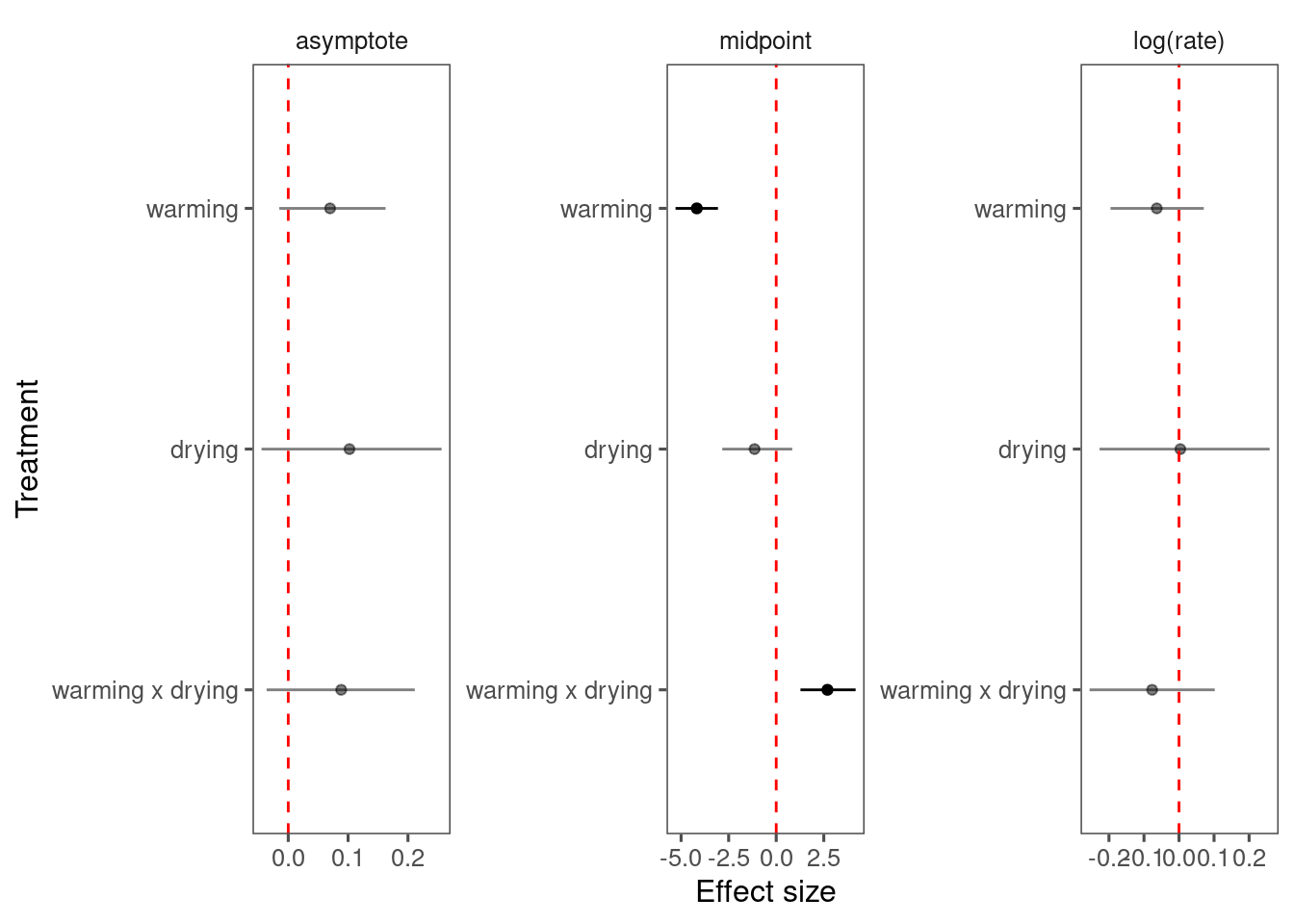
Mixing is way better than Model 4 and Model 5.
Make predictions
df_pred_6 <- calc_bayes_predict(
data = dat_6 %>%
distinct(year, group, heat_trt, water_trt, doy),
df_MCMC = df_MCMC_6,
version = 6
)
write_rds(df_pred_6, "alldata/intermediate/shootmodeling/df_pred_6.rds")
df_pred_6 <- read_rds("alldata/intermediate/shootmodeling/df_pred_6.rds")
p_bayes_predict <- plot_bayes_predict(
data = dat_6,
data_predict = df_pred_6,
vis_log = T
)
p_bayes_predict$p_original
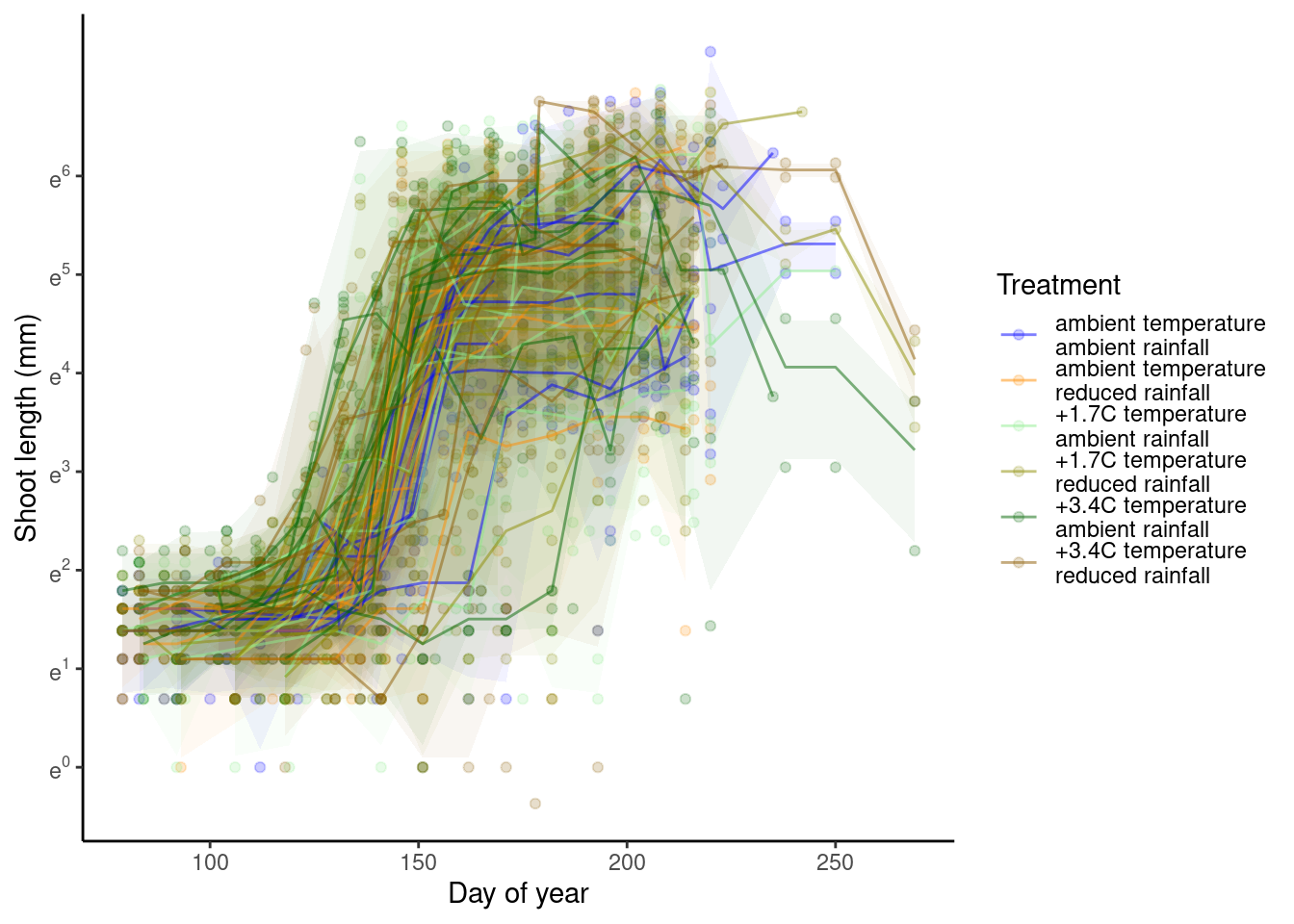
p_bayes_predict$p_overlay
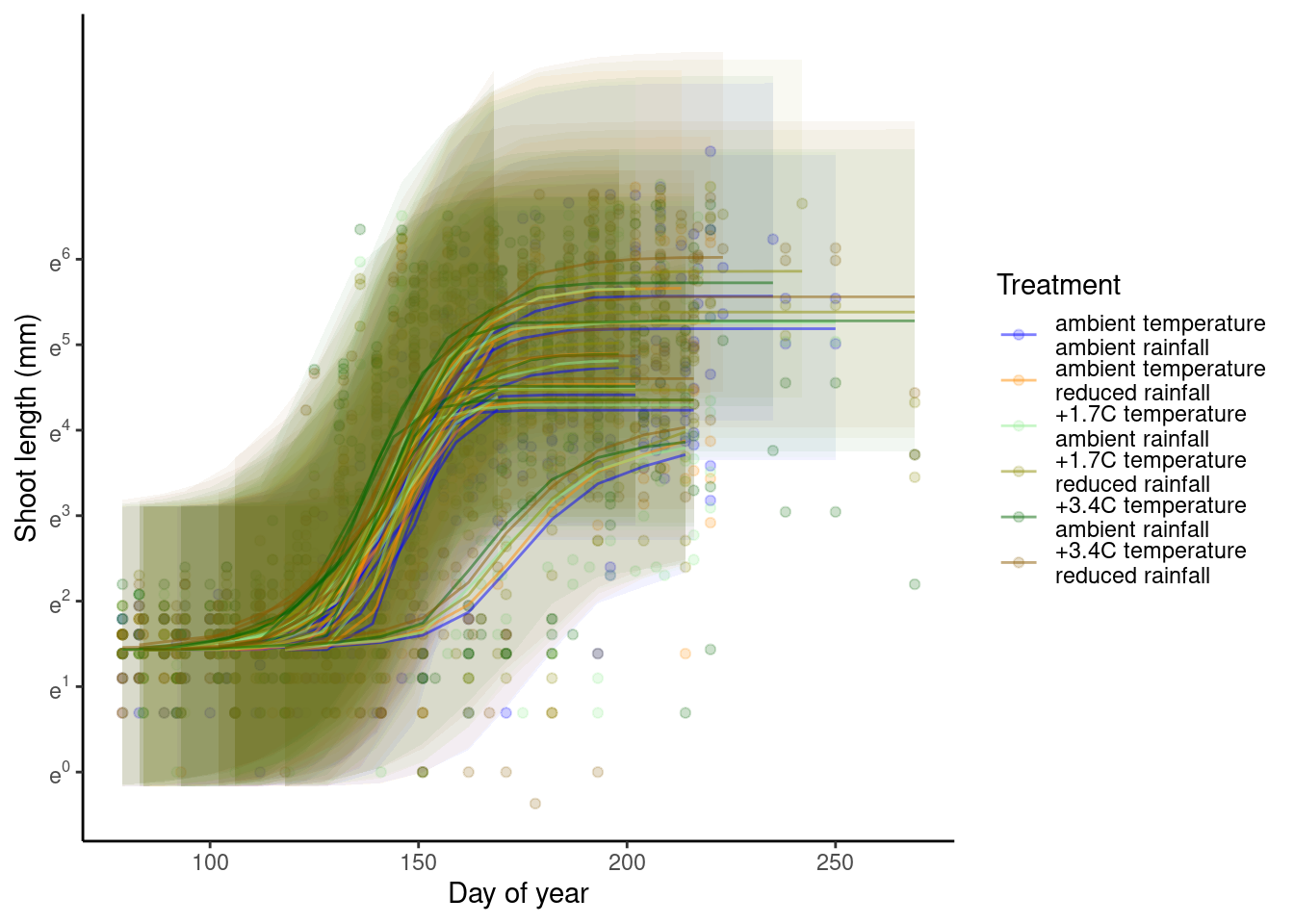
p_bayes_predict$p_accuracy
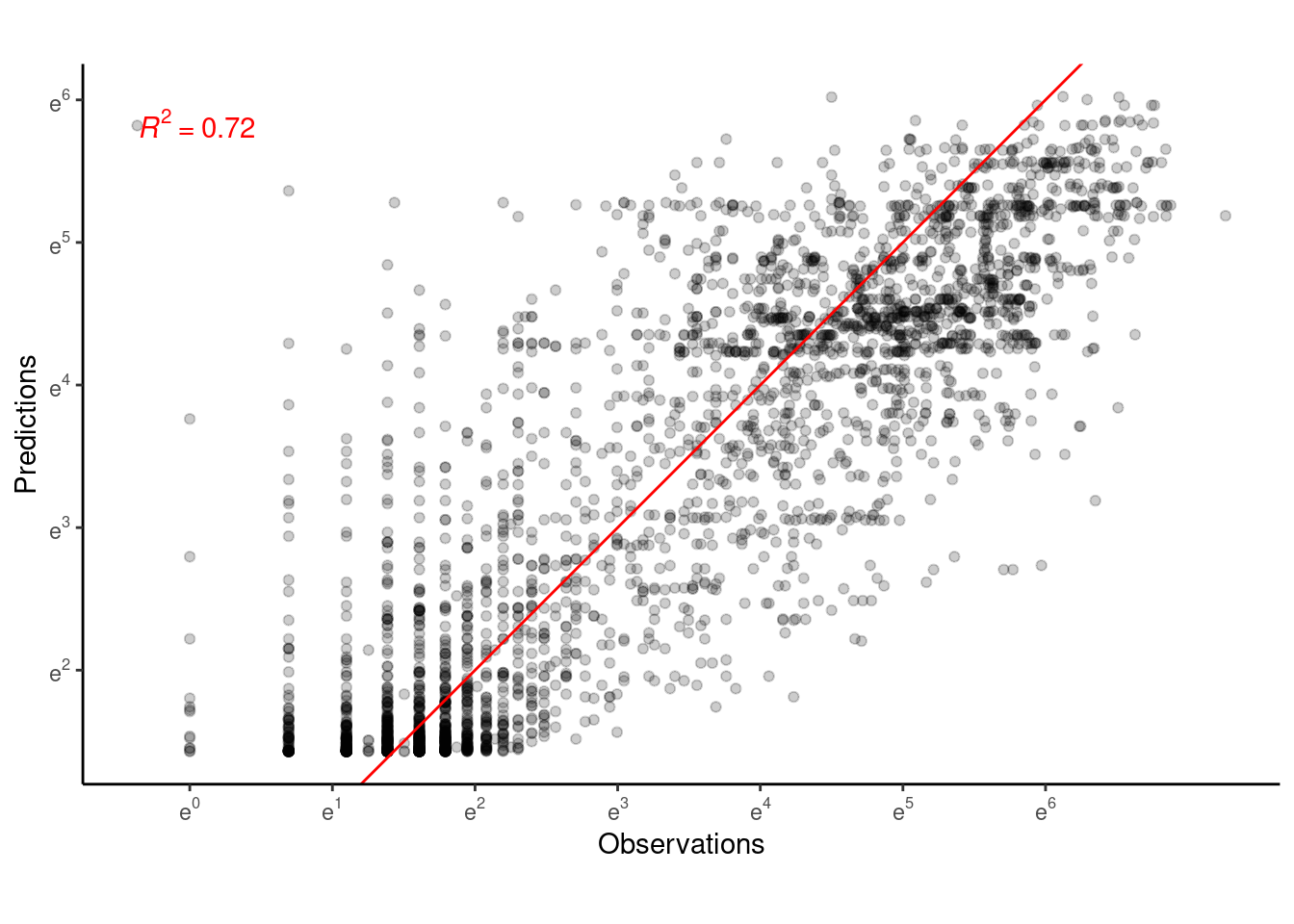
Prediction is not much worse than Model 5.
p_bayes_predict <- plot_bayes_predict(
data = dat_6 %>% filter(year == 2013),
data_predict = df_pred_6 %>% filter(year == 2013),
vis_log = T
)
p_bayes_predict$p_original
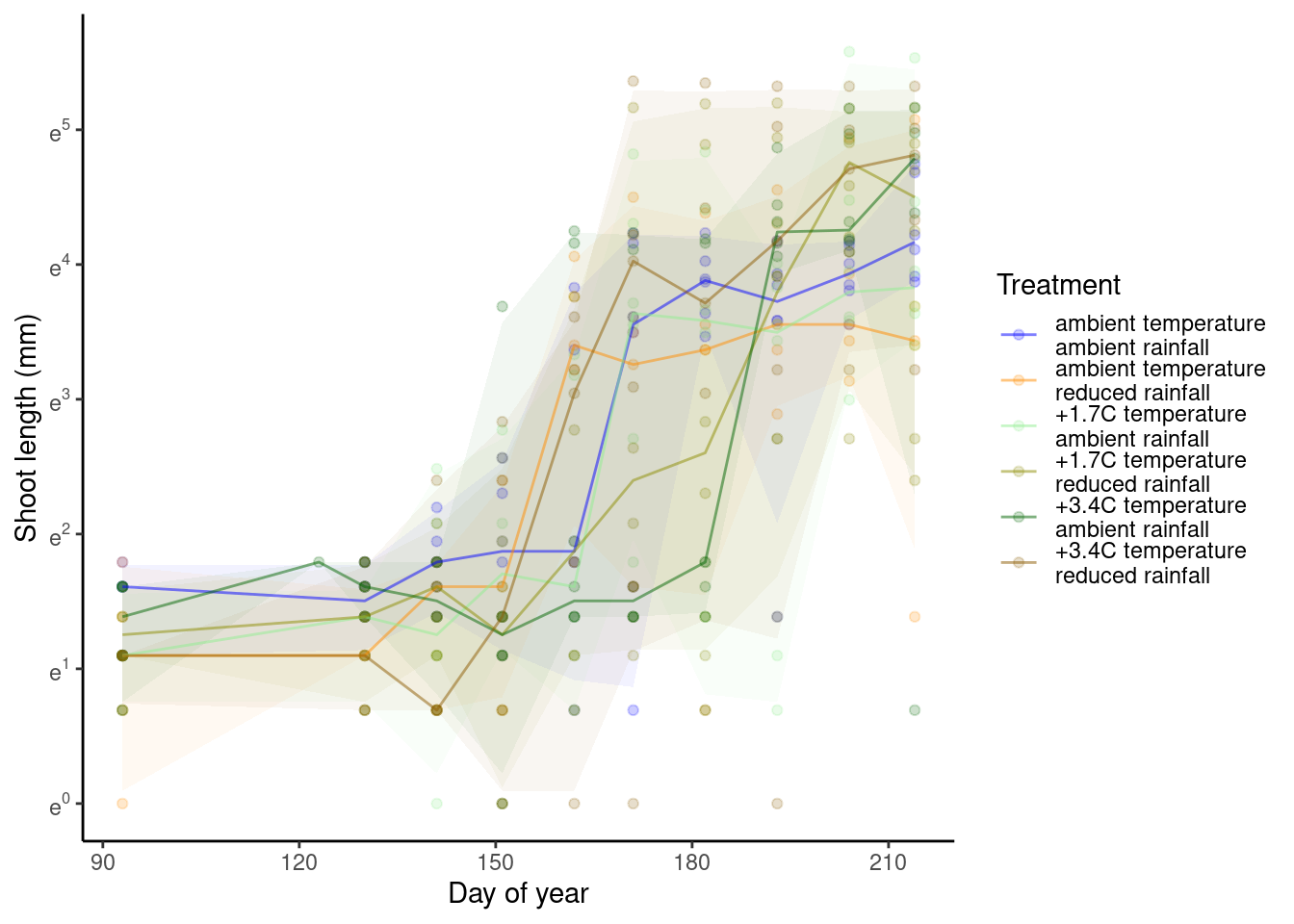
p_bayes_predict$p_overlay
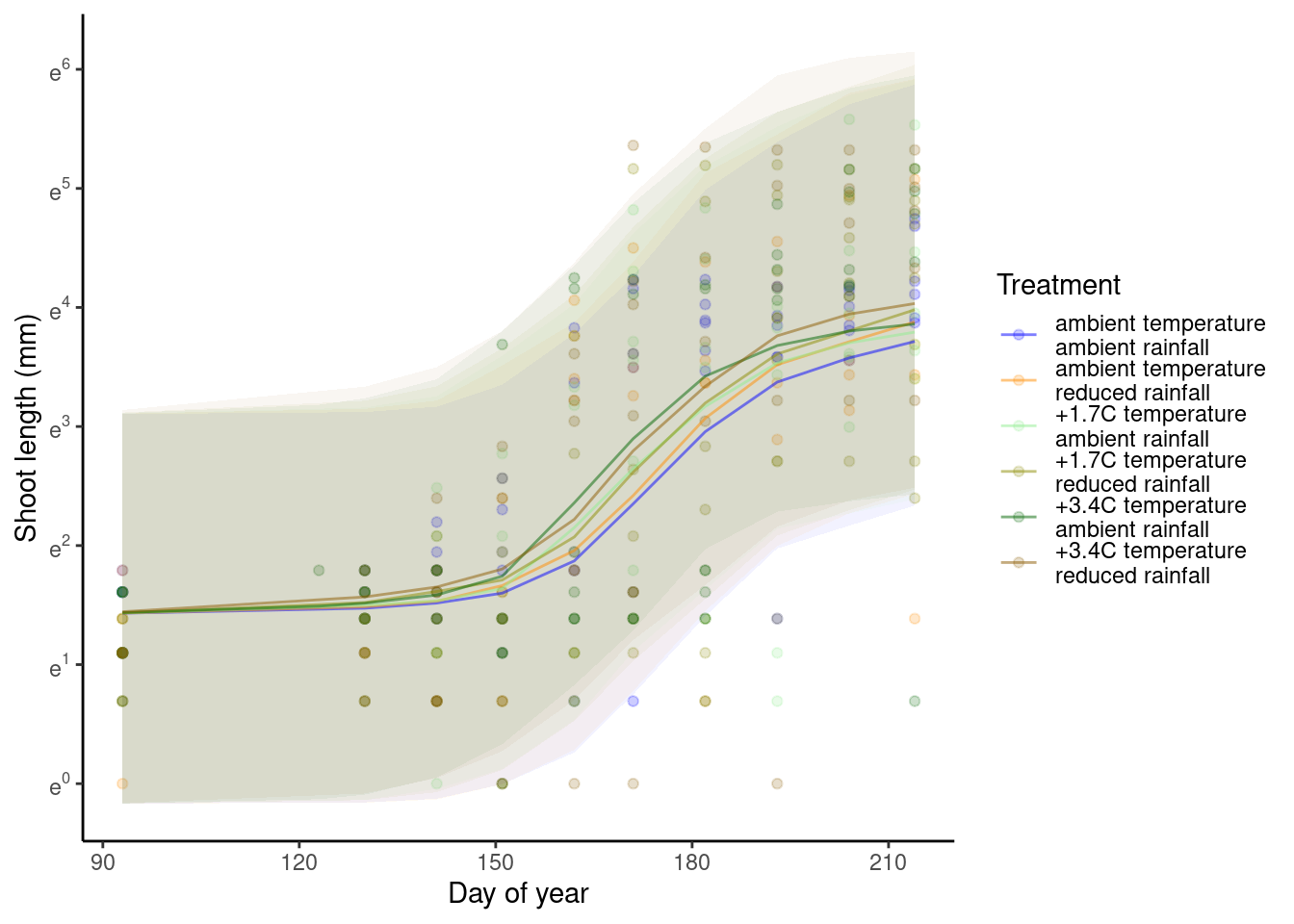 Note that the effects of treatments might be reversed in year 2013, which are not captured by the current model.
Note that the effects of treatments might be reversed in year 2013, which are not captured by the current model.
If you wonder if this is because 2013 was a particularly cold year. I checked that it was colder than others, but it was similar to 2014, which did not have the reversal in treatment effect.
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1)
dat_climate_spring %>%
ggplot() +
geom_point(aes(x = year, y = temp))
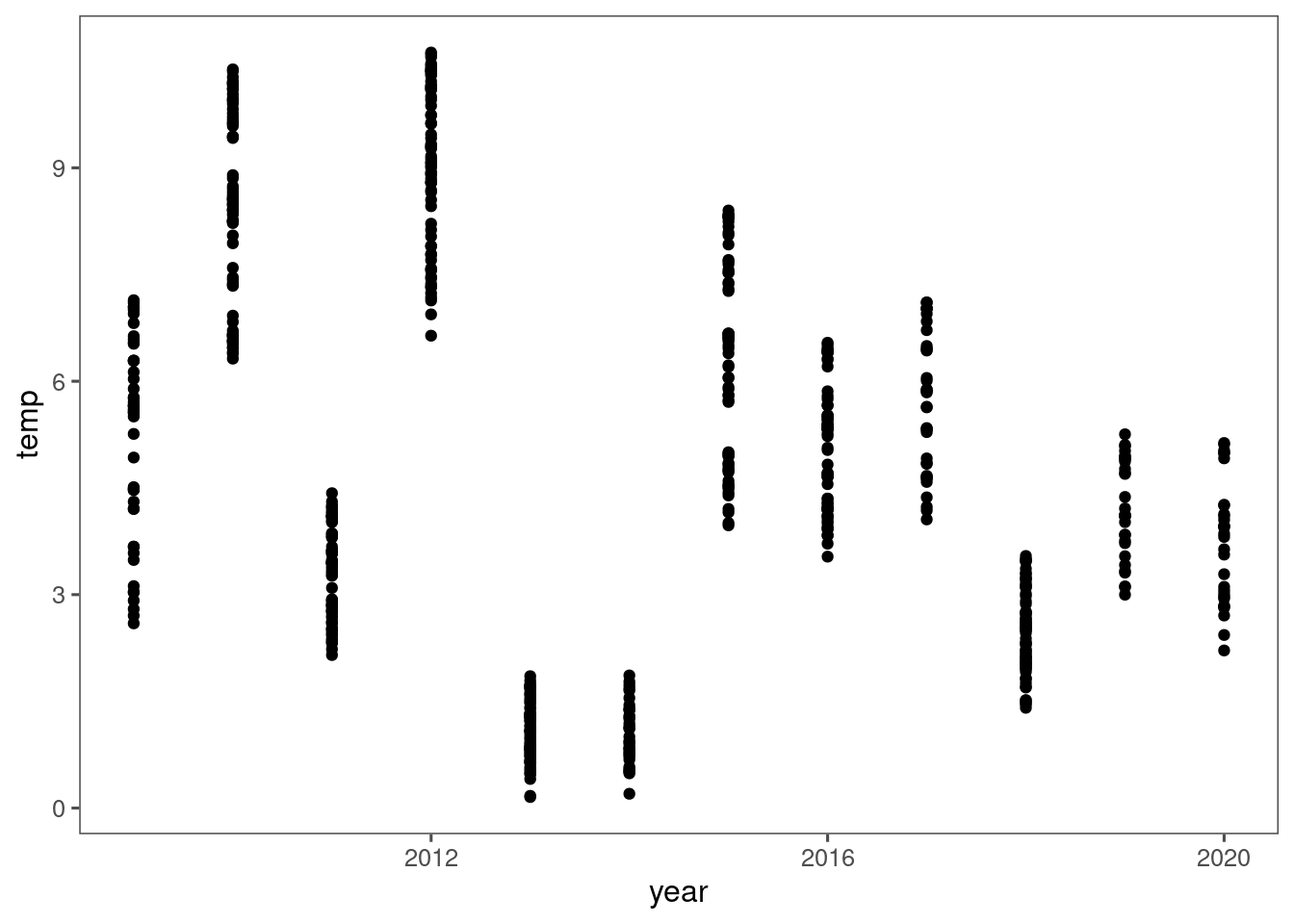 Therefore, I remind us that the inference from this Bayesian model represent an overall effect, which may not be the case for individual years.
Therefore, I remind us that the inference from this Bayesian model represent an overall effect, which may not be the case for individual years.
Bayesian model 7
Shoot-level random effects plus year-level random effects. Treatments as covariates.
Model
Data model
\begin{align*} y_{i,t,d} \sim \text{Lognormal}(\mu_{i,t,d}, \sigma^2) \end{align*}
Process model
\begin{align*} \mu_{i,t,d} &= c+\frac{A_{i,t}}{1+e^{-k_{i,t}(d-x_{0 i,t})}} \newline A_{i,t} &= \mu_A + \delta_{A,i}+ \alpha_{A,i} + \gamma_{A, t}\newline x_{0 i,t} &= \mu_{x_0} + \delta_{x_0,i}+ \alpha_{x_0,i} + \gamma_{x_0,t} \newline log(k_{i,t}) &= \mu_{log(k)} + \delta_{log(k),i}+ \alpha_{log(k),i} + \gamma_{log(k),t} \end{align*}
Fixed effects
\begin{align*} \delta_{A,i} &= \beta_{A,1} T_i + \beta_{A,2} D_i + \beta_{A,3} T_i D_i \newline \delta_{x_0,i} &= \beta_{x_0,1} T_i + \beta_{x_0,2} D_i + \beta_{x_0,3} T_i D_i \newline \delta_{log(k),i} &= \beta_{log(k),1} T_i + \beta_{log(k),2} D_i + \beta_{log(k),3} T_i D_i \end{align*}
Random effects
\begin{align*} \alpha_{A,i} &\sim \text{Normal}(0, \sigma_{A,\alpha}^2) \newline \alpha_{x_0,i} &\sim \text{Normal}(0, \sigma_{x_0,\alpha}^2) \newline \alpha_{log(k),i} &\sim \text{Normal}(0, \sigma_{log(k),\alpha}^2) \newline
\gamma_{A,t} &\sim \text{Normal}(0, \sigma_{A,\gamma}^2) \newline \gamma_{x_0,t} &\sim \text{Normal}(0, \sigma_{x_0,\gamma}^2) \newline \gamma_{log(k),t} &\sim \text{Normal}(0, \sigma_{log(k),\gamma}^2) \end{align*}
Priors
\begin{align*} c &\sim \text{Normal}(0, 1) \newline \mu_A &\sim \text{Normal}(5, 1) \newline \beta_A &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 1 & 0 & 0 \newline 0 & 1 & 0 \newline 0 & 0 & 1 \newline \end{pmatrix} )\newline \sigma_{A,\alpha}^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \sigma_{A,\gamma}^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \mu_{x_0} &\sim \text{Normal}(160, 100) \newline \beta_{x_0} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 100 & 0 & 0 \newline 0 & 100 & 0 \newline 0 & 0 & 100 \newline \end{pmatrix} )\newline \sigma_{x_0, \alpha}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \sigma_{x_0, \gamma}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \mu_{log(k)} &\sim \text{Normal}(-2, 0.04) \newline \beta_{log(k)} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 0.04 & 0 & 0 \newline 0 & 0.04 & 0 \newline 0 & 0 & 0.04 \newline \end{pmatrix} )\newline \sigma_{log(k), \alpha}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma_{log(k), \gamma}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \end{align*}
Prepare data
dat_7 <- dat_shoot_cov %>%
filter(species == "queru", canopy == "open", site == "cfc") %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = str_c(barcode, slot, sep = "_") %>% factor() %>% as.integer()) %>%
mutate(year_code = year %>% factor() %>% as.integer()) %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer())
Fit model
df_MCMC_7 <- calc_bayes_fit(
data = dat_7 %>% mutate(tag = "training"),
version = 7
)
write_rds(df_MCMC_7, "alldata/intermediate/shootmodeling/df_MCMC_7.rds")
df_MCMC_7 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_7.rds") %>% tidy_mcmc(dat_7)
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_7, plot_corr = F)
p_bayes_diagnostics$p_MCMC
p_bayes_diagnostics$p_posterior
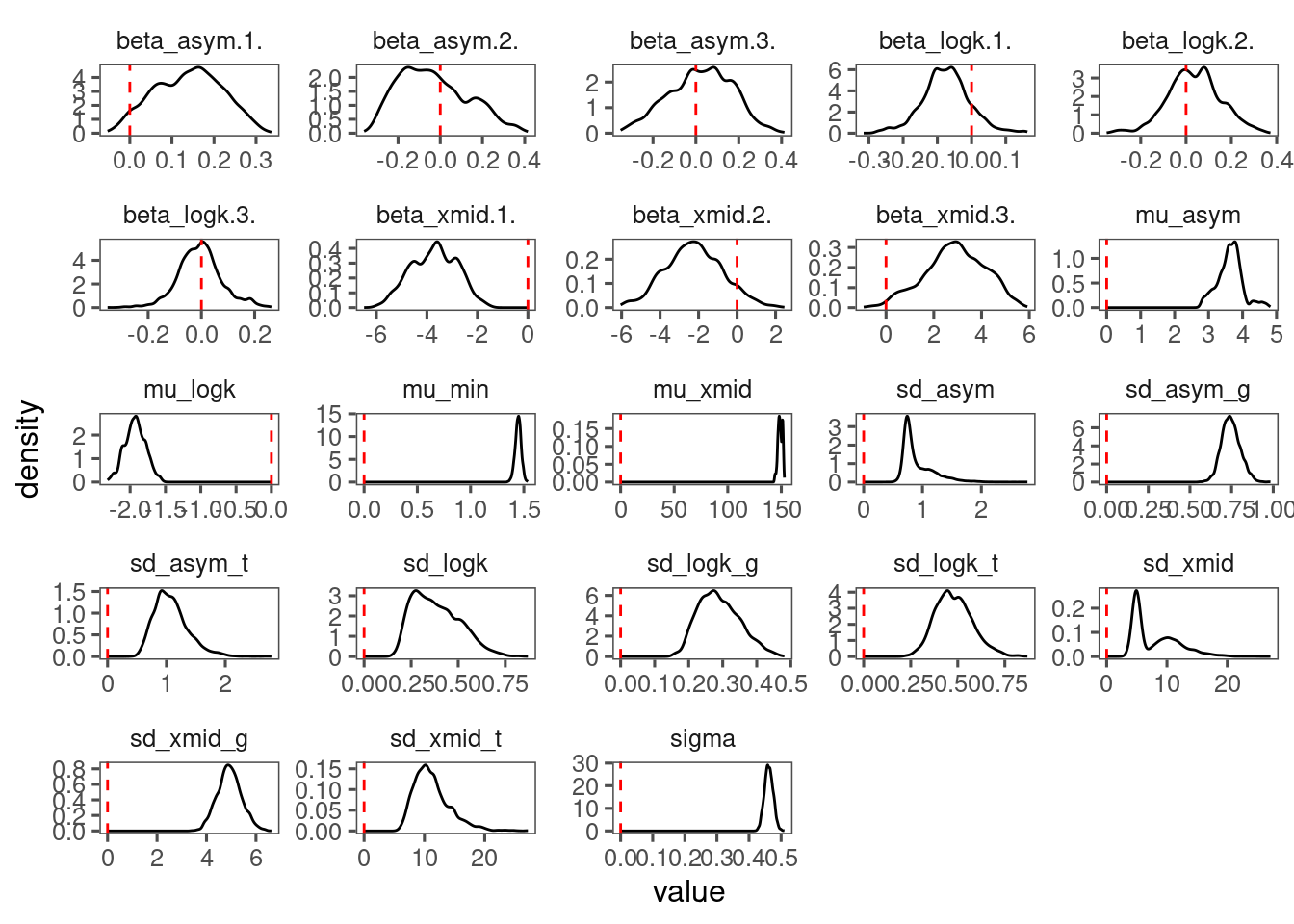
p_bayes_diagnostics$p_coefficient
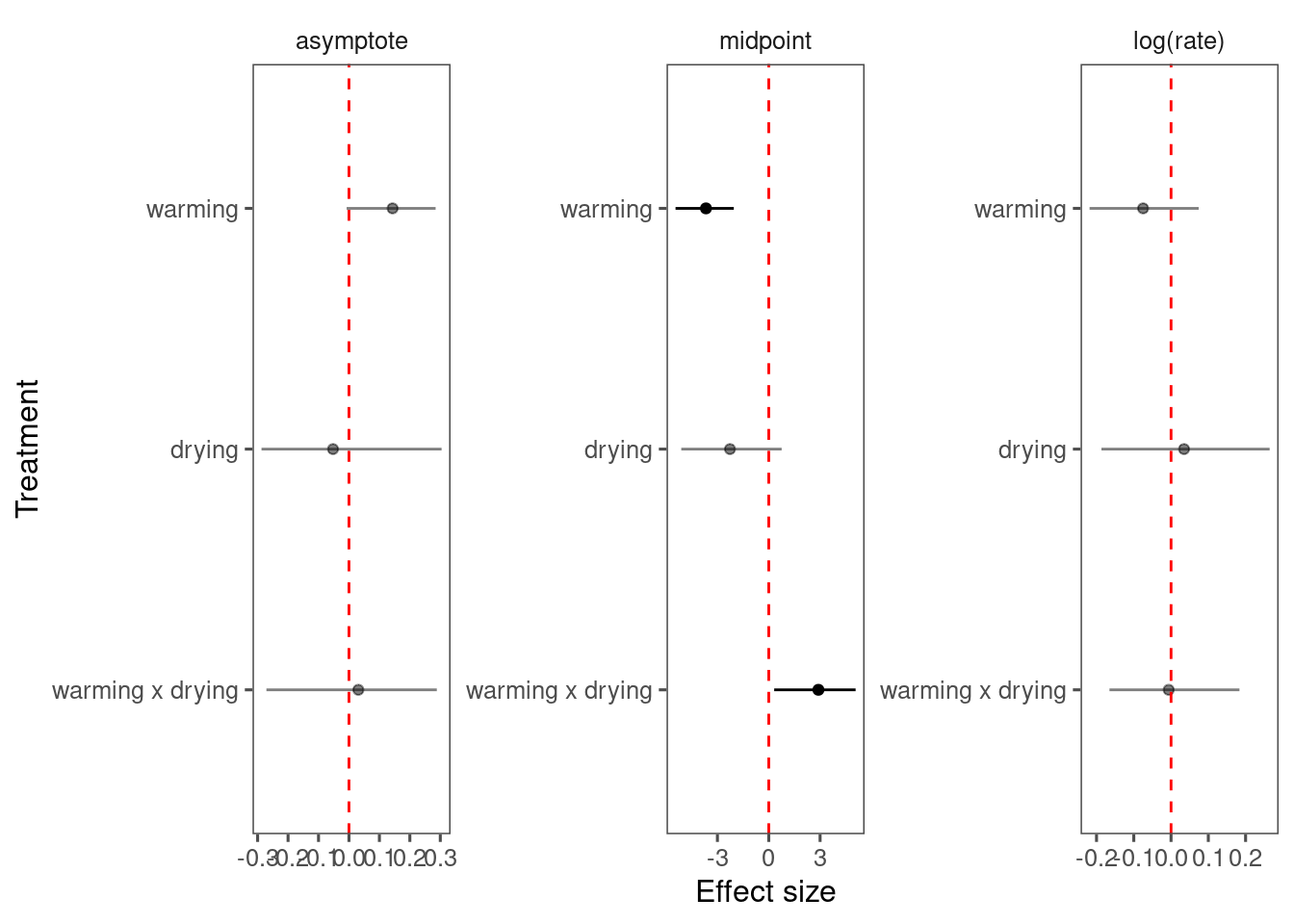
Mixing is not great. Inference similar.
Make predictions
df_pred_7 <- calc_bayes_predict(
data = dat_7 %>%
distinct(group, year, year_code, heat_trt, water_trt, doy),
df_MCMC = df_MCMC_7,
version = 7
)
write_rds(df_pred_7, "alldata/intermediate/shootmodeling/df_pred_7.rds")
df_pred_7 <- read_rds("alldata/intermediate/shootmodeling/df_pred_7.rds")
p_bayes_predict <- plot_bayes_predict(
data = dat_7,
data_predict = df_pred_7,
vis_log = T
)
p_bayes_predict$p_original
p_bayes_predict$p_overlay
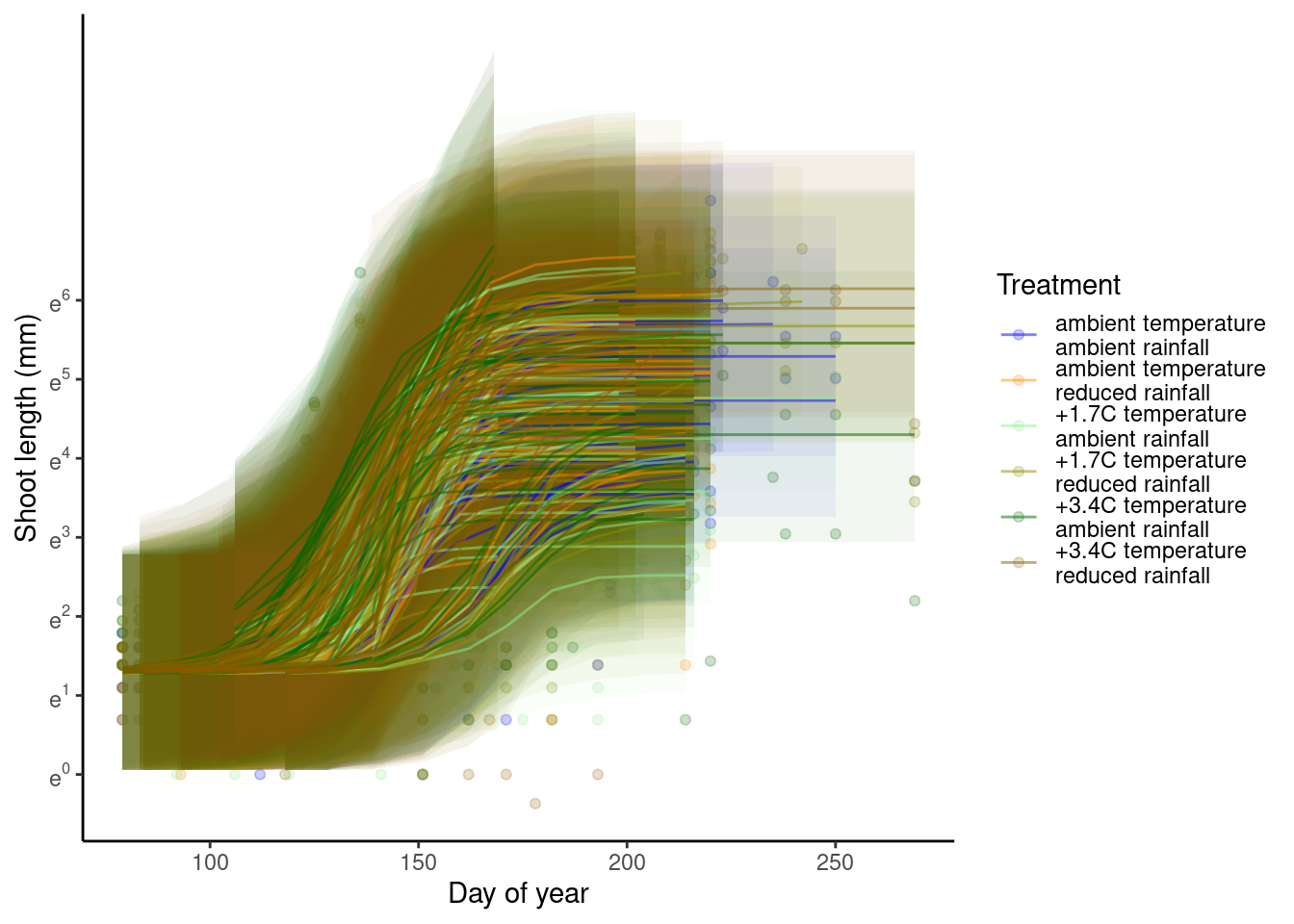
p_bayes_predict$p_accuracy
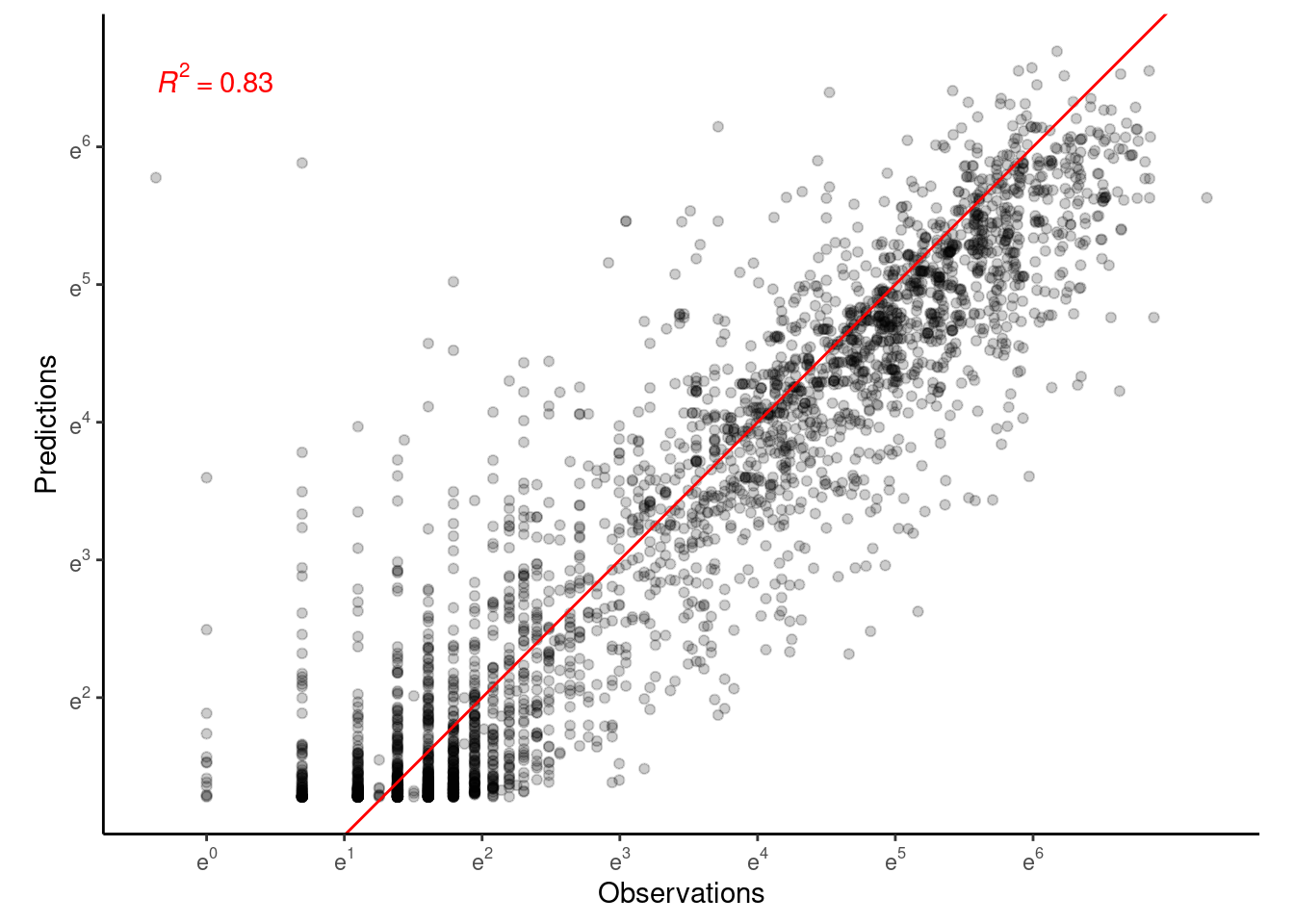
Fit not too bad compared to Model 4.
Bayesian model 8
Shoot-year-level random effects with temporally autoregressive parameters over years for each shoot. Treatments as covariates. This is written but not recommended, because I think inter-annual variations are greater than variations between shoots within a year. It might be hard to describe the effects of warm vs. cold years with temporal autoregression.
Model
Data model
\begin{align*} y_{i,t,d} \sim \text{Lognormal}(\mu_{i,t,d}, \sigma^2) \end{align*}
Process model
\begin{align*} \mu_{i,t,d} &= c+\frac{A_{i,t}}{1+e^{-k_{i}(d-x_{0 i,t})}} \newline A_{i,t} &= \mu_A + \delta_{A,i}+ \alpha_{A,i,t} \newline x_{0 i,t} &= \mu_{x_0} + \delta_{x_0, i}+ \alpha_{x_0,i,t} \newline log(k_{i,t}) &= \mu_{log(k)}+ \delta_{log(k),i}+ \alpha_{log(k),i,t} \end{align*}
Fixed effects
\begin{align*} \delta_{A,i} &= \beta_{A,1} T_i + \beta_{A,2} D_i + \beta_{A,3} T_i D_i \newline \delta_{x_0,i} &= \beta_{x_0,1} T_i + \beta_{x_0,2} D_i + \beta_{x_0,3} T_i D_i \newline \delta_{log(k),i} &= \beta_{log(k),1} T_i + \beta_{log(k),2} D_i + \beta_{log(k),3} T_i D_i \end{align*}
Random effects
\begin{align*} \alpha_{A,i,1} &\sim \text{Normal}(0, \sigma_{A,1}^2) \newline \alpha_{A,i,t} &\sim \text{Normal}(\alpha_{A,i,t-1}, \sigma_{A,t}^2) \newline \alpha_{x_0,i,1} &\sim \text{Normal}(0,\sigma_{x_0,1}^2) \newline \alpha_{x_0,i,t} &\sim \text{Normal}(\alpha_{x_0,i,t-1}, \sigma_{x_0,t}^2) \newline \alpha_{log(k),i,1} &\sim \text{Normal}(0,\sigma_{log(k),1}^2) \newline \alpha_{log(k),i,t} &\sim \text{Normal}(\alpha_{log(k),i,t-1}, \sigma_{log(k),t}^2) \end{align*}
Priors
\begin{align*} c &\sim \text{Normal}(0, 1) \newline \mu_A &\sim \text{Normal}(5, 1) \newline \beta_A &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 1 & 0 & 0 \newline 0 & 1 & 0 \newline 0 & 0 & 1 \newline \end{pmatrix} )\newline \sigma_{A,1}^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \sigma_{A,t}^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \newline \mu_{x_0} &\sim \text{Normal}(160, 100) \newline \beta_{x_0} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 100 & 0 & 0 \newline 0 & 100 & 0 \newline 0 & 0 & 100 \newline \end{pmatrix} )\newline \sigma_{x_0,1}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \sigma_{x_0,t}^2 &\sim \text{Truncated Normal}(0, 100, 0, \infty) \newline \mu_{log(k)} &\sim \text{Normal}(-2, 0.04) \newline \beta_{log(k)} &\sim \text{Multivariate Normal} ( \begin{pmatrix} 0 \newline 0 \newline 0 \newline \end{pmatrix}, \begin{pmatrix} 0.04 & 0 & 0 \newline 0 & 0.04 & 0 \newline 0 & 0 & 0.04 \newline \end{pmatrix} )\newline \sigma_{log(k),1}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma_{log(k),t}^2 &\sim \text{Truncated Normal}(0, 0.04, 0, \infty) \newline \sigma^2 &\sim \text{Truncated Normal}(0, 1, 0, \infty) \end{align*}
Prepare data
dat_8 <- dat_shoot_cov %>%
filter(species == "queru", canopy == "open", site == "cfc") %>%
filter(shoot > 0) %>%
drop_na(barcode) %>%
mutate(slot = if_else(is.na(slot), "", slot)) %>%
mutate(group = str_c(barcode, slot, sep = "_") %>% factor() %>% as.integer()) %>%
group_by(group) %>%
mutate(year_code = year %>% factor() %>% as.integer()) %>%
ungroup() %>%
mutate(heat_trt = factor(heat_name, levels = c("ambient", "+1.7C", "+3.4C"), labels = c(0, 1, 2)) %>% as.character() %>% as.integer()) %>%
mutate(water_trt = factor(water_name, levels = c("ambient", "reduced"), labels = c(0, 1)) %>% as.character() %>% as.integer())
Fit model
df_MCMC_8 <- calc_bayes_fit(
data = dat_8 %>% mutate(tag = "training"),
version = 8
)
write_rds(df_MCMC_8, "alldata/intermediate/shootmodeling/df_MCMC_8.rds")
df_MCMC_8 <- read_rds("alldata/intermediate/shootmodeling/df_MCMC_8.rds") %>% tidy_mcmc(dat_8)
p_bayes_diagnostics <- plot_bayes_diagnostics(df_MCMC = df_MCMC_8, plot_corr = F)
p_bayes_diagnostics$p_MCMC
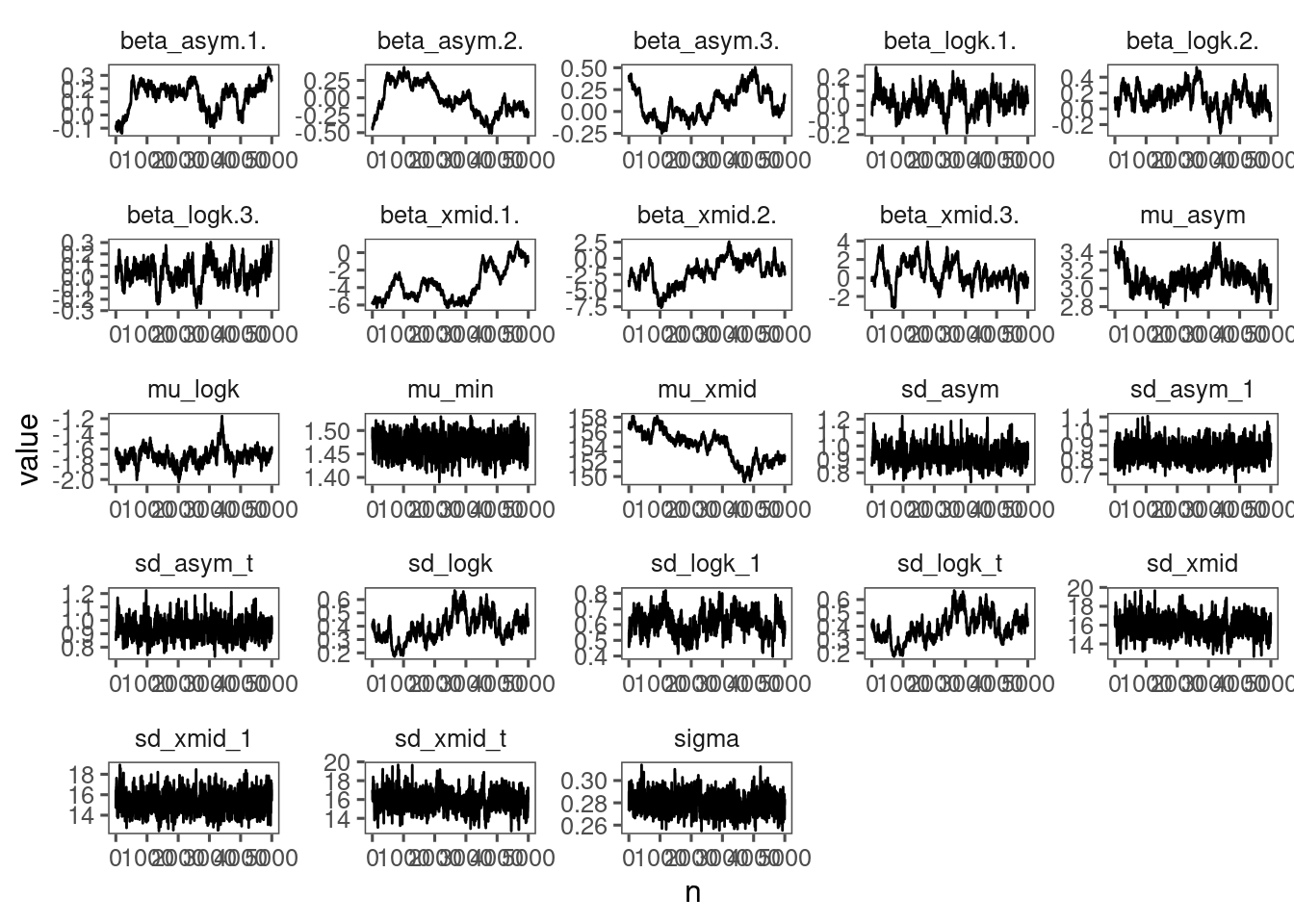
p_bayes_diagnostics$p_posterior
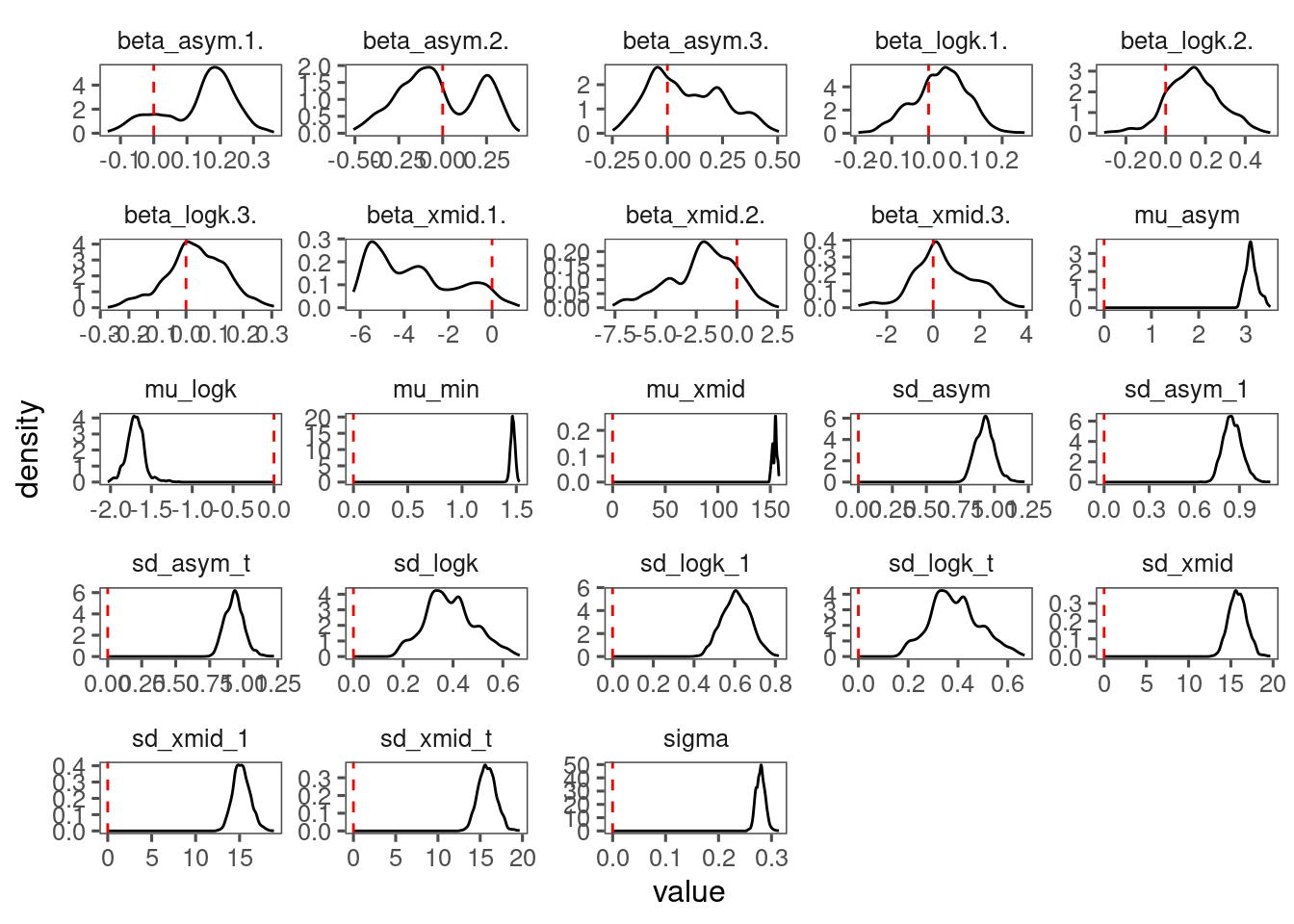
p_bayes_diagnostics$p_coefficient
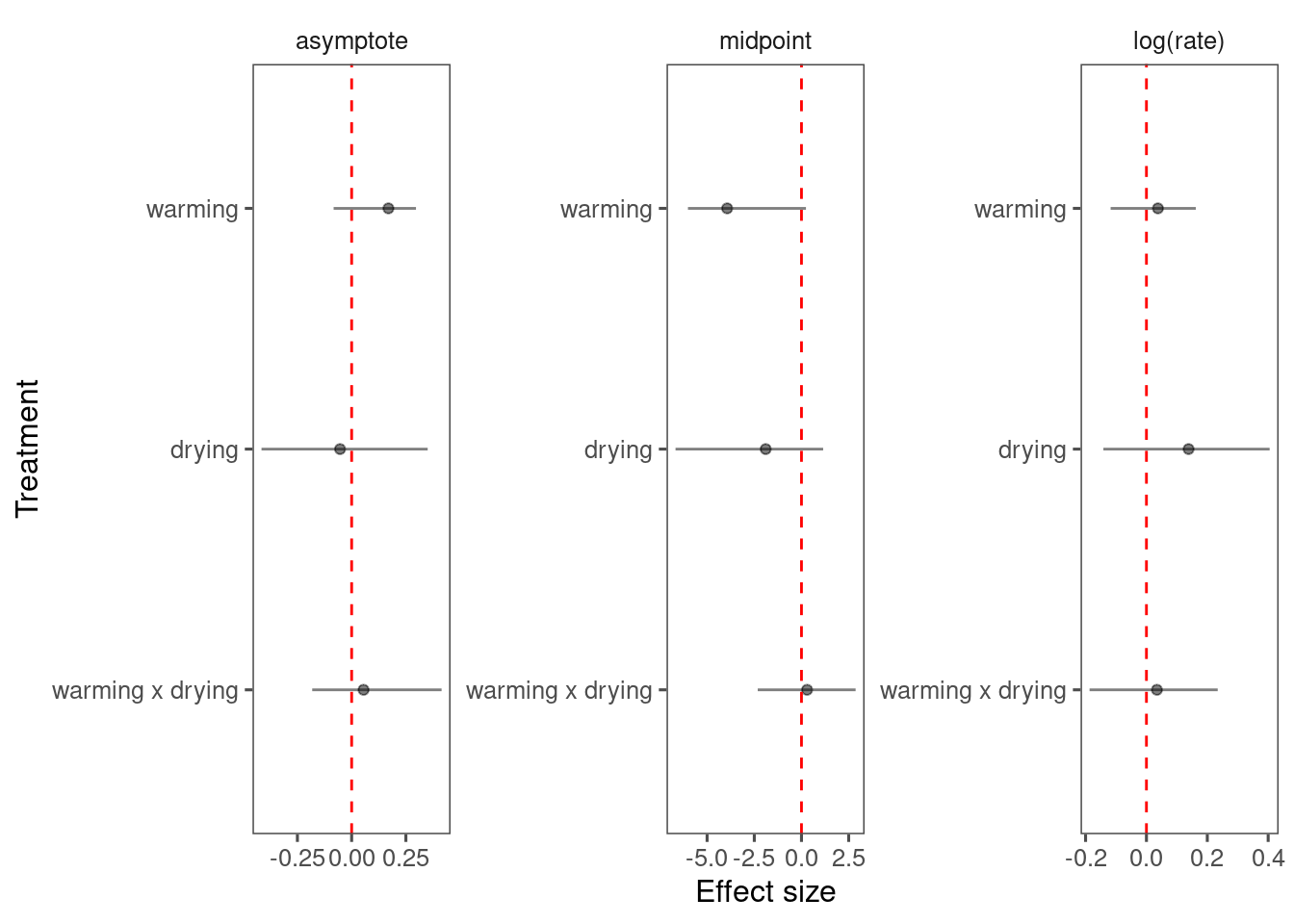 Mixing is as bad as Model 4.
Mixing is as bad as Model 4.
Make predictions
df_pred_8 <- calc_bayes_predict(
data = dat_8 %>%
distinct(group, year, year_code, heat_trt, water_trt, doy),
df_MCMC = df_MCMC_8,
version = 8
)
write_rds(df_pred_8, "alldata/intermediate/shootmodeling/df_pred_8.rds")
df_pred_8 <- read_rds("alldata/intermediate/shootmodeling/df_pred_8.rds")
p_bayes_predict <- plot_bayes_predict(
data = dat_8,
data_predict = df_pred_8,
vis_log = T
)
p_bayes_predict$p_original
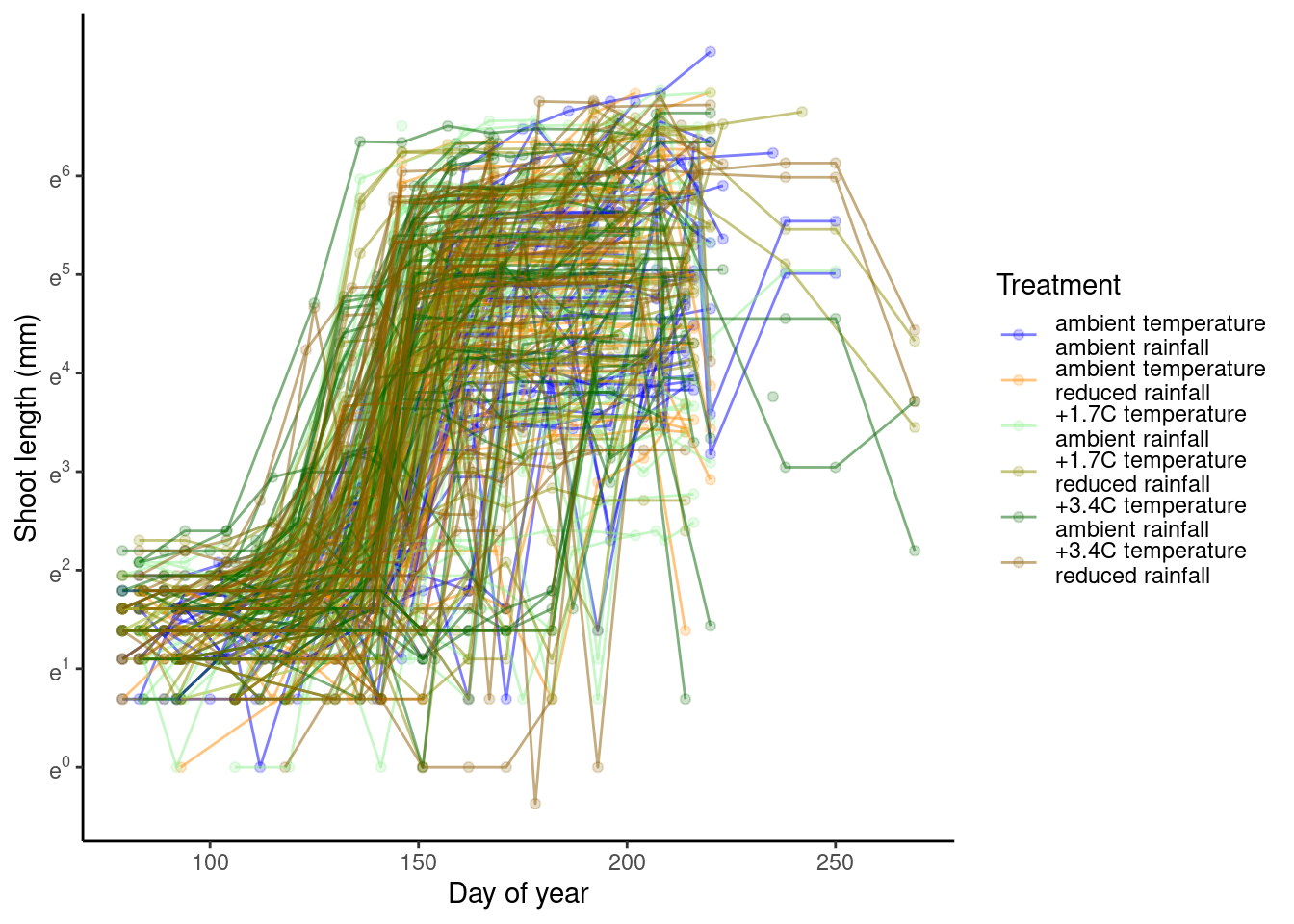
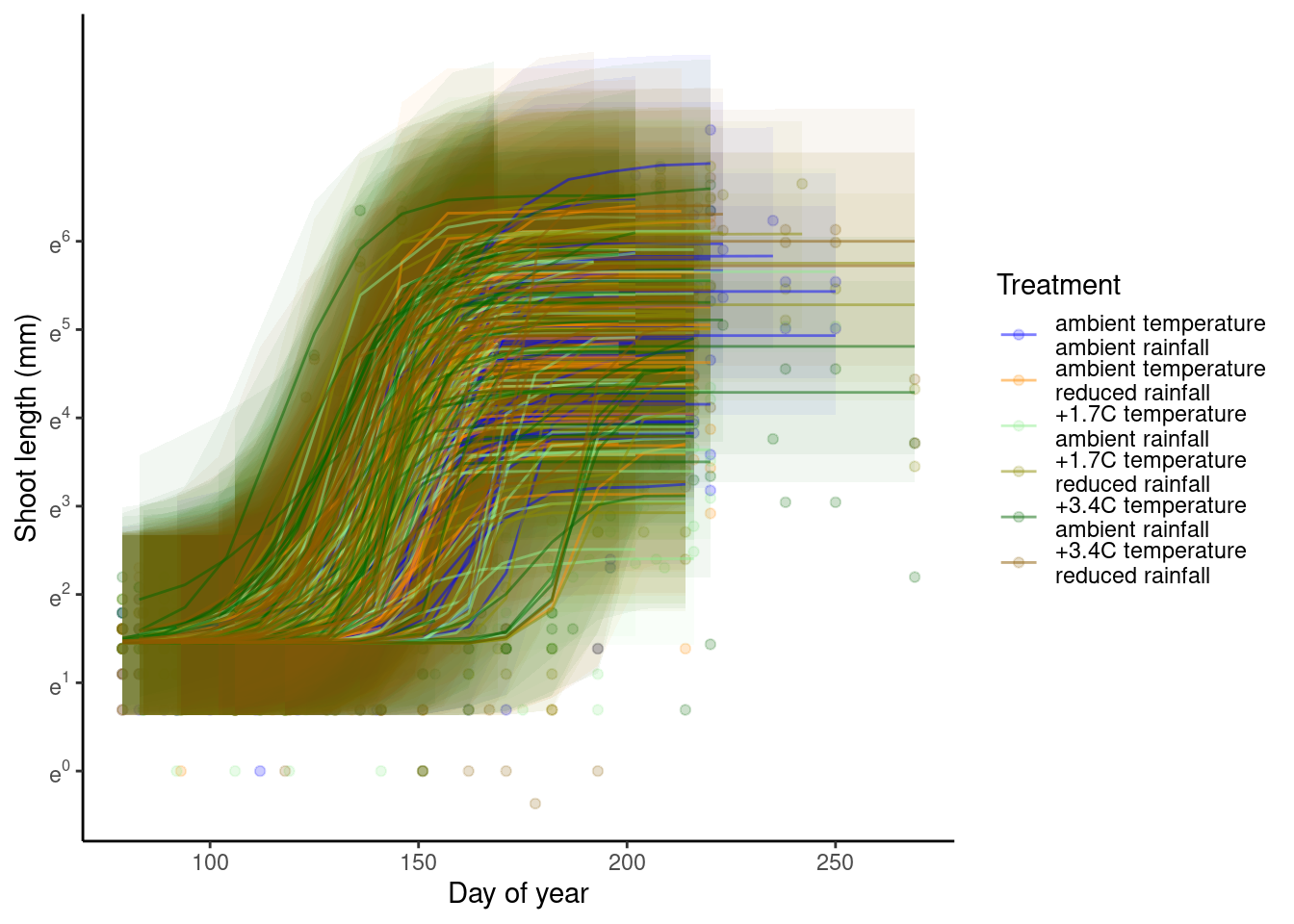
p_bayes_predict$p_overlay
p_bayes_predict$p_accuracy
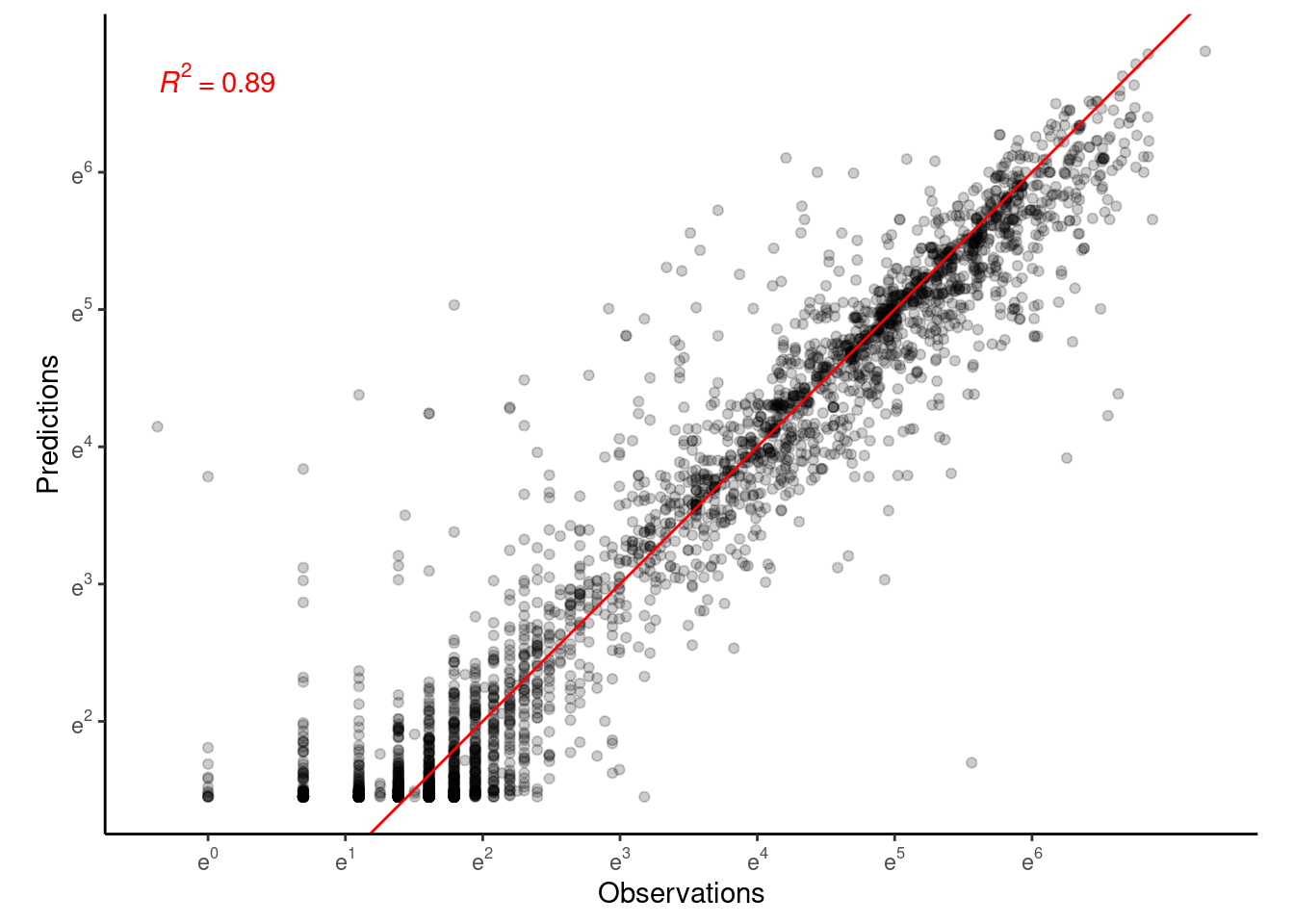 Performance similar to Model 4.
Performance similar to Model 4.