Treatment effects on start of phenophase
dat_phenophase## # A tibble: 4,116,485 × 16
## site canopy heat heat_name water water_name block plot species barcode
## <chr> <fct> <fct> <fct> <fct> <fct> <chr> <chr> <chr> <dbl>
## 1 cfc open _ ambient _ ambient d d1 abiba 1
## 2 cfc open _ ambient _ ambient d d1 abiba 1
## 3 cfc open _ ambient _ ambient d d1 abiba 1
## 4 cfc open _ ambient _ ambient d d1 abiba 1
## 5 cfc open _ ambient _ ambient d d1 abiba 1
## 6 cfc open _ ambient _ ambient d d1 abiba 1
## 7 cfc open _ ambient _ ambient d d1 abiba 1
## 8 cfc open _ ambient _ ambient d d1 abiba 1
## 9 cfc open _ ambient _ ambient d d1 abiba 1
## 10 cfc open _ ambient _ ambient d d1 abiba 1
## # ℹ 4,116,475 more rows
## # ℹ 6 more variables: cohort <dbl>, year <dbl>, doy <dbl>, phenophase <fct>,
## # status <dbl>, notes <chr>dat_phenophase_time <- calc_phenophase_time(dat_phenophase)
dat_phenophase_time$phenophase %>% unique()## [1] budbreak oneleaf mostleaf senescence leafdrop
## Levels: budbreak oneleaf mostleaf senescence leafdropdat_phenophase_diff <- calc_phenophase_diff(dat_phenophase_time)p_phenophase_diff <- plot_phenophase_diff(dat_phenophase_diff %>%
filter(species %in% c("queru", "aceru")) %>%
mutate(species = factor(species,
levels = c("queru", "aceru"),
labels = c("Red oak", "Red maple")
)))
p_phenophase_diff$budbreak## NULLggsave(filename = "alldata/output/figures/budbreak_effect.png", p_phenophase_diff$budbreak)
p_phenophase_diff$senescence## NULLggsave(filename = "alldata/output/figures/senescence_effect.png", p_phenophase_diff$senescence)Statistical test with LME.
mod <- lmerTest::lmer(diff ~ heat_name * water_name + (1 | species), data = dat_phenophase_diff %>%
filter(phenophase == "budbreak"))
summary(mod)## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: diff ~ heat_name * water_name + (1 | species)
## Data: dat_phenophase_diff %>% filter(phenophase == "budbreak")
##
## REML criterion at convergence: 148383.1
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -7.4515 -0.2989 -0.0160 0.2232 12.4937
##
## Random effects:
## Groups Name Variance Std.Dev.
## species (Intercept) 1.09 1.044
## Residual 203.52 14.266
## Number of obs: 18195, groups: species, 21
##
## Fixed effects:
## Estimate Std. Error df t value
## (Intercept) -8.183e-02 3.159e-01 2.780e+01 -0.259
## heat_name+1.7 °C -3.406e+00 2.854e-01 1.817e+04 -11.932
## heat_name+3.4 °C -5.815e+00 2.875e-01 1.817e+04 -20.225
## water_namereduced -1.450e-01 4.691e-01 1.783e+04 -0.309
## heat_name+1.7 °C:water_namereduced 4.825e-01 6.632e-01 1.816e+04 0.728
## heat_name+3.4 °C:water_namereduced 9.474e-01 6.570e-01 1.817e+04 1.442
## Pr(>|t|)
## (Intercept) 0.798
## heat_name+1.7 °C <2e-16 ***
## heat_name+3.4 °C <2e-16 ***
## water_namereduced 0.757
## heat_name+1.7 °C:water_namereduced 0.467
## heat_name+3.4 °C:water_namereduced 0.149
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) ht_+17°C ht_+34°C wtr_nm h_+17°C:
## het_nm+17°C -0.440
## het_nm+34°C -0.438 0.483
## watr_nmrdcd -0.264 0.296 0.293
## ht_n+17°C:_ 0.190 -0.430 -0.208 -0.701
## ht_n+34°C:_ 0.192 -0.211 -0.438 -0.708 0.501mod <- lmerTest::lmer(diff ~ heat_name * water_name + (1 | species), data = dat_phenophase_diff %>%
filter(phenophase == "senescence"))
summary(mod)## Linear mixed model fit by REML. t-tests use Satterthwaite's method [
## lmerModLmerTest]
## Formula: diff ~ heat_name * water_name + (1 | species)
## Data: dat_phenophase_diff %>% filter(phenophase == "senescence")
##
## REML criterion at convergence: 130236.3
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -11.6581 -0.4996 -0.0098 0.5083 5.7334
##
## Random effects:
## Groups Name Variance Std.Dev.
## species (Intercept) 4.872 2.207
## Residual 110.035 10.490
## Number of obs: 17267, groups: species, 22
##
## Fixed effects:
## Estimate Std. Error df t value
## (Intercept) -1.651e-01 5.115e-01 2.326e+01 -0.323
## heat_name+1.7 °C 3.424e+00 2.145e-01 1.724e+04 15.962
## heat_name+3.4 °C 5.097e+00 2.160e-01 1.724e+04 23.596
## water_namereduced 4.578e-02 3.588e-01 1.726e+04 0.128
## heat_name+1.7 °C:water_namereduced 2.422e-01 5.057e-01 1.724e+04 0.479
## heat_name+3.4 °C:water_namereduced -1.081e+00 5.005e-01 1.724e+04 -2.161
## Pr(>|t|)
## (Intercept) 0.7497
## heat_name+1.7 °C <2e-16 ***
## heat_name+3.4 °C <2e-16 ***
## water_namereduced 0.8985
## heat_name+1.7 °C:water_namereduced 0.6320
## heat_name+3.4 °C:water_namereduced 0.0307 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation of Fixed Effects:
## (Intr) ht_+17°C ht_+34°C wtr_nm h_+17°C:
## het_nm+17°C -0.201
## het_nm+34°C -0.200 0.476
## watr_nmrdcd -0.121 0.287 0.285
## ht_n+17°C:_ 0.086 -0.424 -0.202 -0.699
## ht_n+34°C:_ 0.088 -0.206 -0.432 -0.707 0.501Load climate data
dat_climate <- read_climate(option = "raw")
dat_climate_daily <- summ_climate_daily(dat_climate)
setwd("phenologyb4warmed/")
usethis::use_data(dat_climate_daily, overwrite = T)
setwd("..")I saved daily climatic data. Use directly.
dat_climate_spring <- summ_climate_season(dat_climate_daily, date_start = "Mar 15", date_end = "May 15", rainfall = 1, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))
dat_climate_fall <- summ_climate_season(dat_climate_daily, date_start = "Jun 15", date_end = "Sep 15", rainfall = 0, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))
dat_climate_annual_early <- summ_climate_season(dat_climate_daily, date_start = "May 16", start_rollback = 1, date_end = "May 15", end_rollback = 0, rainfall = 0.6, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))
dat_climate_annual_late <- summ_climate_season(dat_climate_daily, date_start = "Sep 16", start_rollback = 1, date_end = "Sep 15", end_rollback = 0, rainfall = 0.6, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "year"))Compare experimental and temporal trends
dat_phenophase_time_summ <- summ_phenophase_time(dat_phenophase_time, group_vars = c("site", "canopy", "heat", "heat_name", "water", "water_name", "species", "phenophase", "year"))Had to remove year 2012 as the phenology looked like outliers given the temperature.
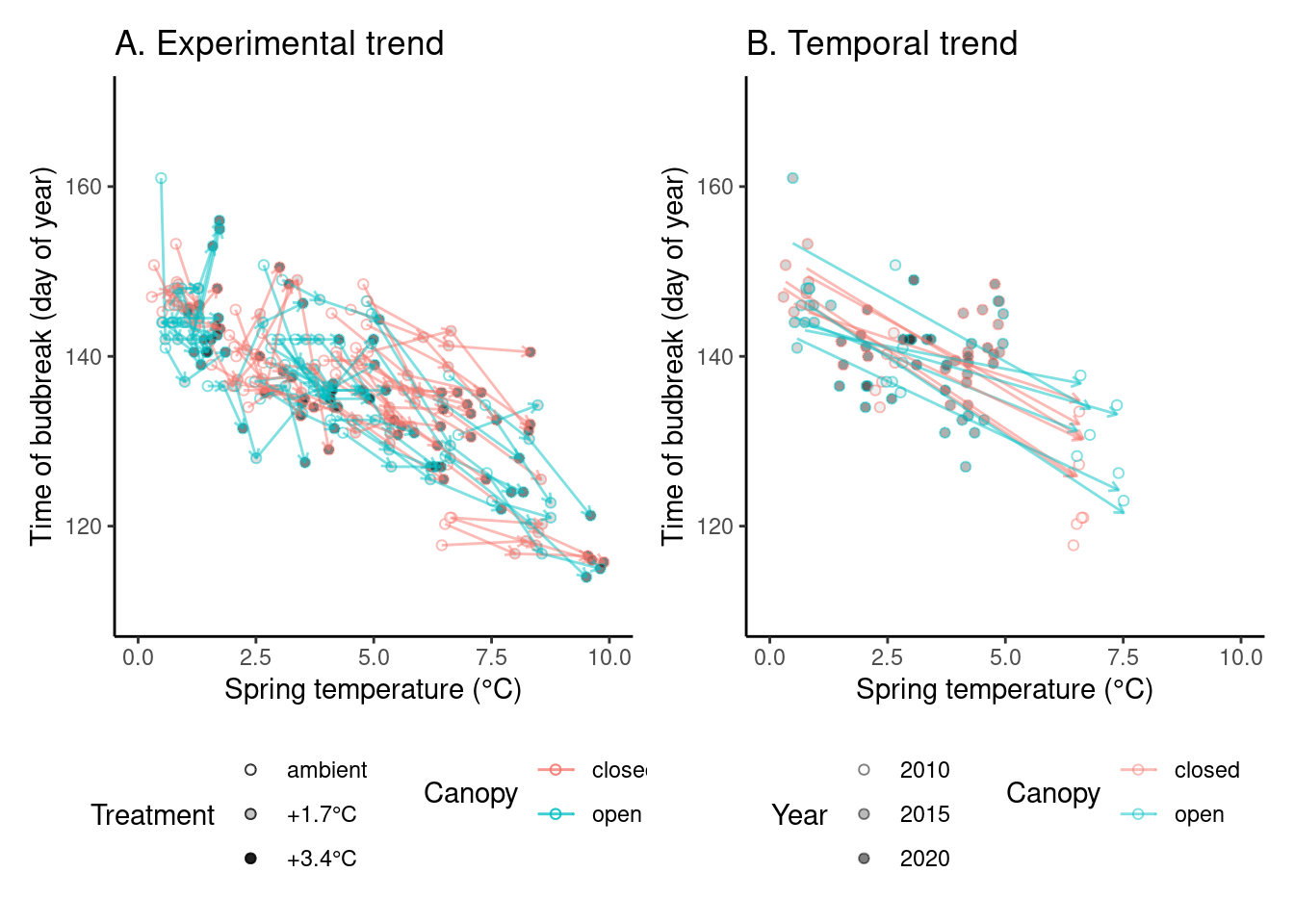
(p_trend1 <- plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "budbreak") %>%
filter(species == "queru") %>%
filter(year != 2012),
dat_climate = dat_climate_spring,
var = "heat"
))
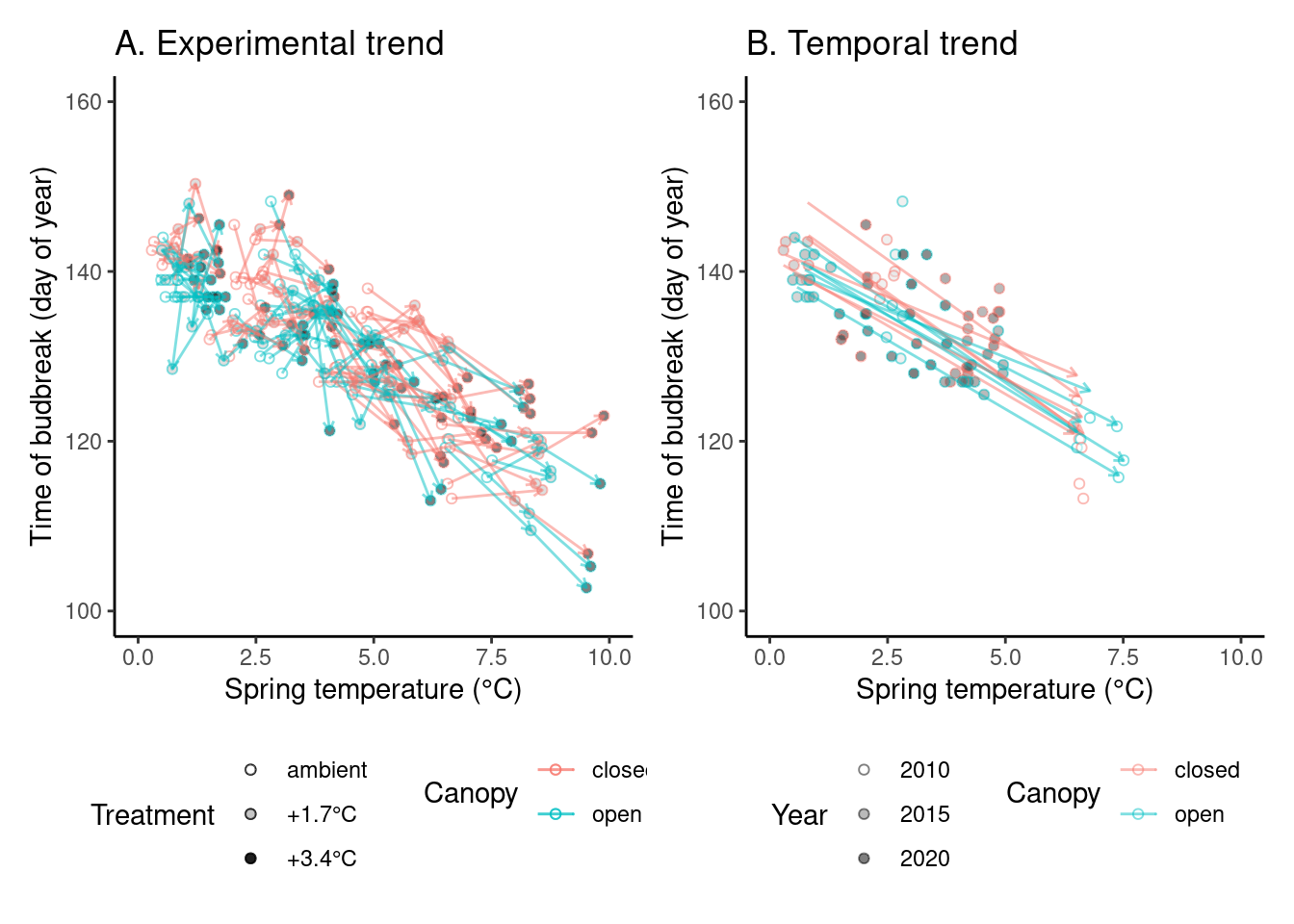
# ggsave("alldata/output/figures/trend_queru.png", p_trend1, width = 12, height = 6)(p_trend2 <- plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "budbreak") %>%
filter(year != 2012),
dat_climate = dat_climate_spring,
var = "heat"
))
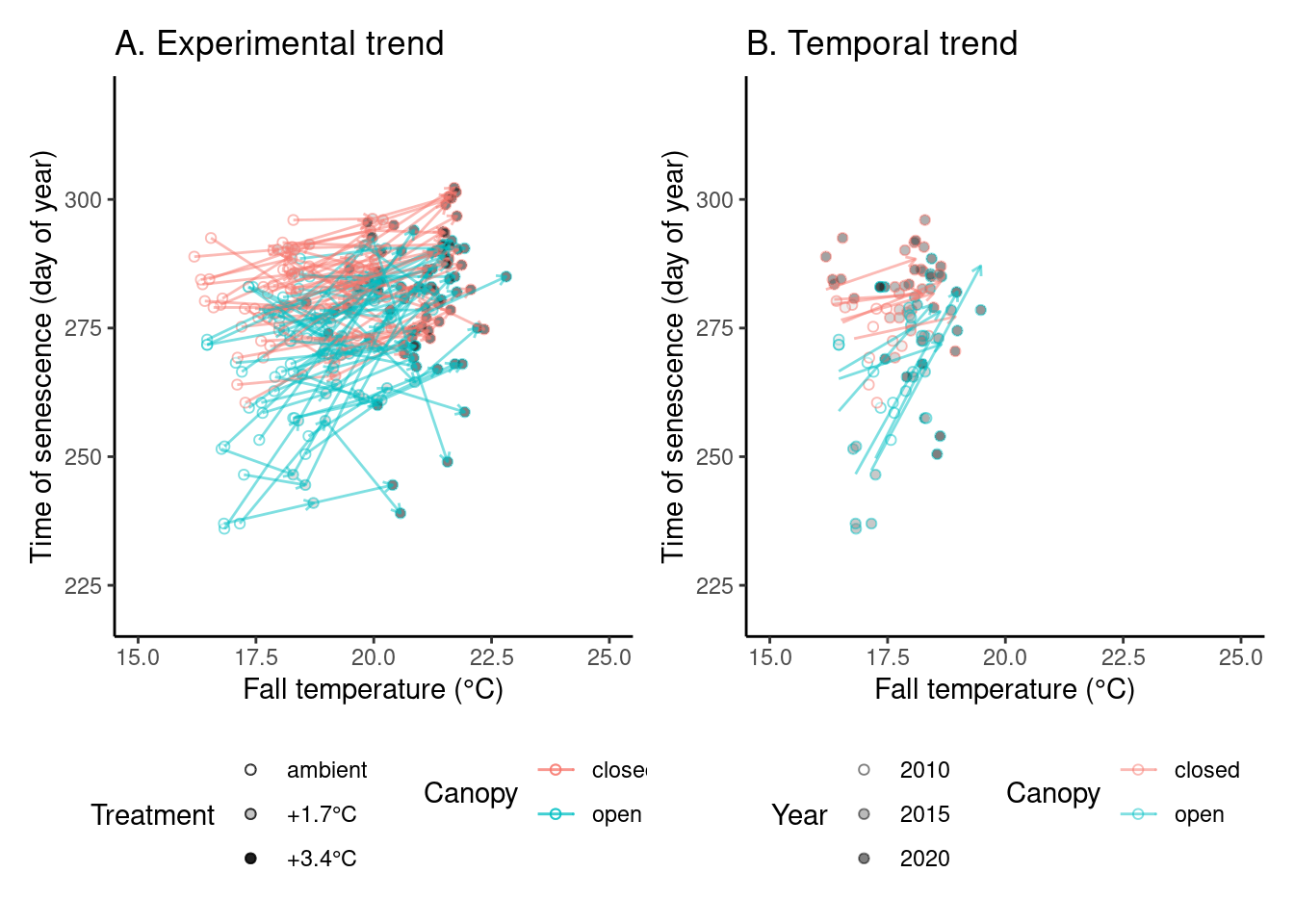
# ggsave("alldata/output/figures/trend_aceru.png", p_trend2, width = 12, height = 6)(p_trend3 <- plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "senescence") %>%
filter(species == "queru") %>%
filter(year != 2012),
dat_climate = dat_climate_fall,
var = "heat"
))
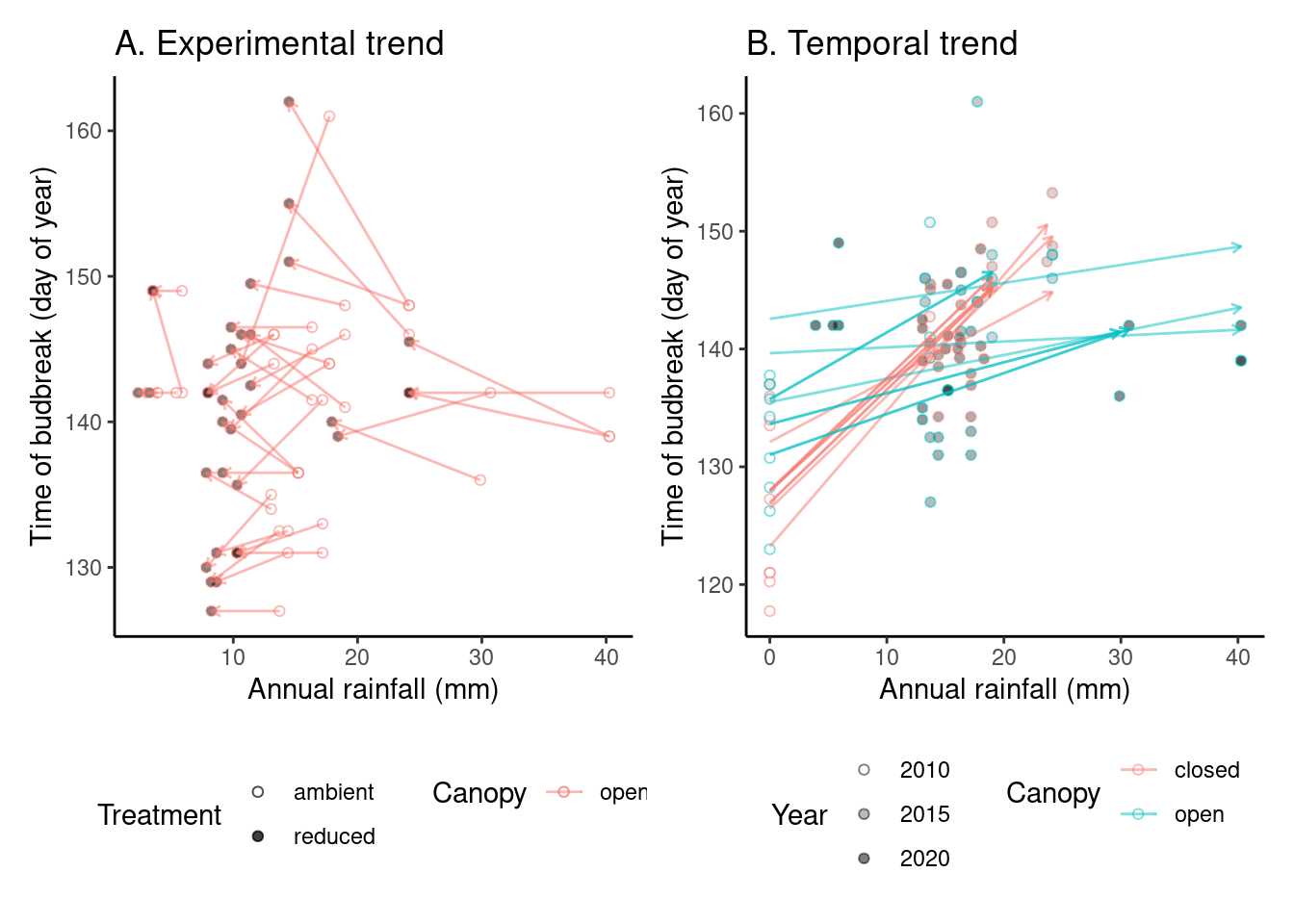
# ggsave("alldata/output/figures/trend_queru_senescence.png", p_trend3, width = 12, height = 6)(p_trend4 <- plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "budbreak") %>%
filter(species == "queru") %>%
filter(year != 2012),
dat_climate = dat_climate_annual_early,
var = "water",
clim_season = "Annual"
))
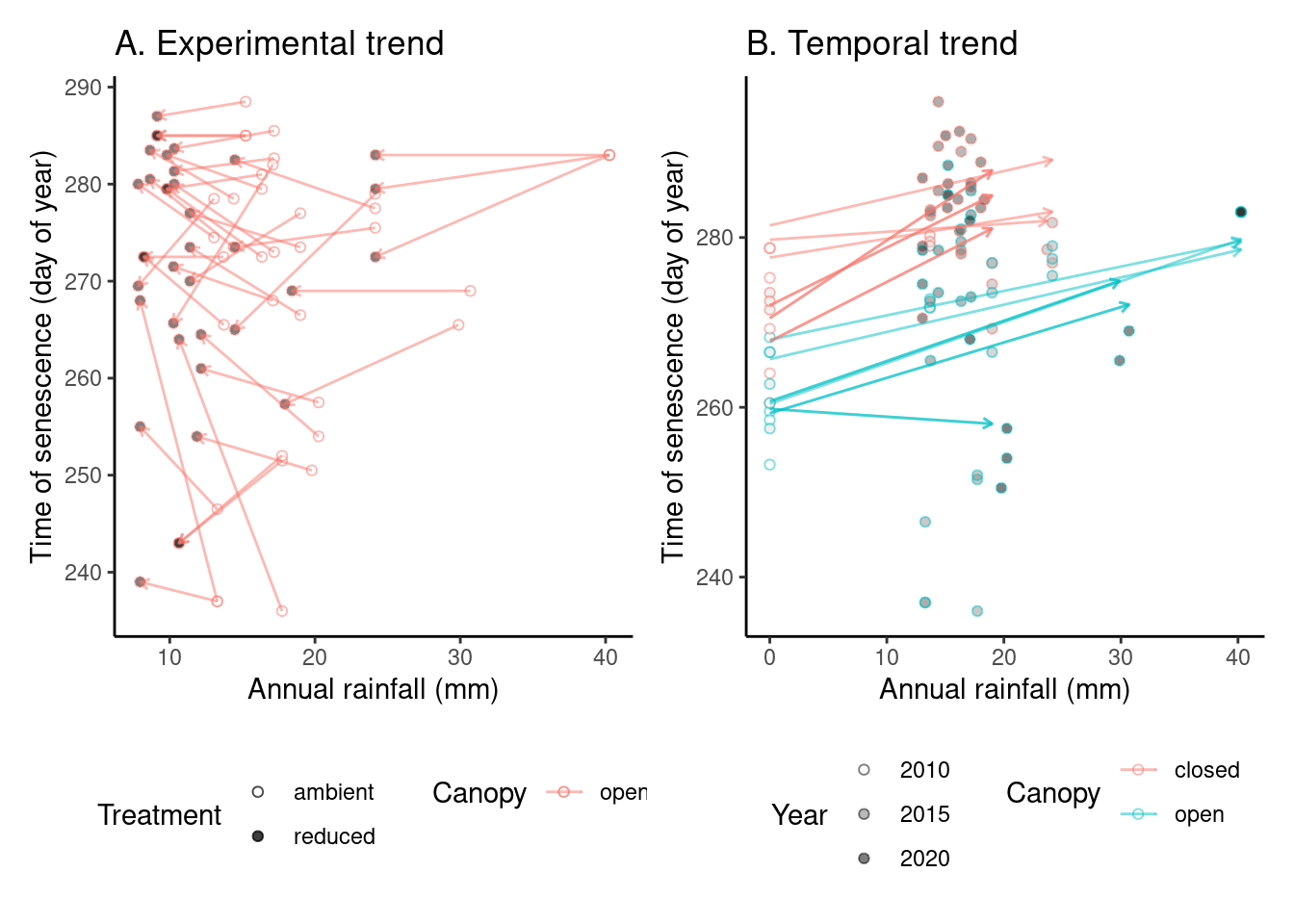
# ggsave("alldata/output/figures/trend_queru_rainfall.png", p_trend4, width = 12, height = 6)(p_trend5 <- plot_trend(
dat_phenophase = dat_phenophase_time_summ %>%
filter(phenophase == "senescence") %>%
filter(species == "queru") %>%
filter(year != 2012),
dat_climate = dat_climate_annual_late,
var = "water",
clim_season = "Annual"
))
# ggsave("alldata/output/figures/trend_queru_rainfall_senescence.png", p_trend5, width = 12, height = 6)